Dual Transcriptomics To Determine Gamma Interferon-Independent Host Response to Intestinal Cryptosporidium parvum Infection
- PMID: 34928716
- PMCID: PMC8852703
- DOI: 10.1128/iai.00638-21
Dual Transcriptomics To Determine Gamma Interferon-Independent Host Response to Intestinal Cryptosporidium parvum Infection
Abstract
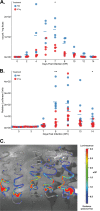
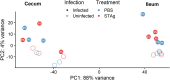
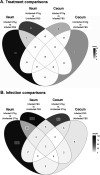
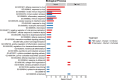
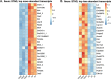
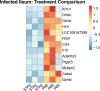
Animals with a chronic infection of the parasite Toxoplasma gondii are protected against lethal secondary infection with other pathogens. Our group previously determined that soluble T. gondii antigens (STAg) can mimic this protection and be used as a treatment against several lethal pathogens. Because treatments are limited for the parasite Cryptosporidium parvum, we tested STAg as a C. parvum therapeutic. We determined that STAg treatment reduced C. parvum Iowa II oocyst shedding in gamma interferon knockout (IFN-γ-KO) mice. Murine intestinal sections were then sequenced to define the IFN-γ-independent transcriptomic response to C. parvum infection. Gene Ontology and transcript abundance comparisons showed host immune response and metabolism changes. Transcripts for type I interferon-responsive genes were more abundant in C. parvum-infected mice treated with STAg. Comparisons between phosphate-buffered saline (PBS) and STAg treatments showed no significant differences in C. parvum gene expression. C. parvum transcript abundance was highest in the ileum and mucin-like glycoproteins and the GDP-fucose transporter were among the most abundant. These results will assist the field in determining both host- and parasite-directed future therapeutic targets.
Keywords: Cryptosporidium parvum; RNA sequencing; STAg; Toxoplasma gondii; cecum; ileum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
MyD88-dependent pathways mediate resistance to Cryptosporidium parvum infection in mice.Infect Immun. 2006 Jan;74(1):549-56. doi: 10.1128/IAI.74.1.549-556.2006. Infect Immun. 2006. PMID: 16369011 Free PMC article.
-
Dynamics of gut mucosal and systemic Th1/Th2 cytokine responses in interferon-gamma and interleukin-12p40 knock out mice during primary and challenge Cryptosporidium parvum infection.Immunobiology. 2009;214(6):454-66. doi: 10.1016/j.imbio.2008.11.015. Epub 2009 Jan 19. Immunobiology. 2009. PMID: 19155092
-
Profiles of healing and nonhealing Cryptosporidium parvum infection in C57BL/6 mice with functional B and T lymphocytes: the extent of gamma interferon modulation determines the outcome of infection.Infect Immun. 1997 Nov;65(11):4761-9. doi: 10.1128/iai.65.11.4761-4769.1997. Infect Immun. 1997. PMID: 9353062 Free PMC article.
-
Innate and cell-mediated immune responses to Cryptosporidium parvum.Adv Parasitol. 1998;40:87-119. doi: 10.1016/s0065-308x(08)60118-9. Adv Parasitol. 1998. PMID: 9554071 Review.
-
The Mucosal Innate Immune Response to Cryptosporidium parvum, a Global One Health Issue.Front Cell Infect Microbiol. 2021 May 25;11:689401. doi: 10.3389/fcimb.2021.689401. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34113580 Free PMC article. Review.
Cited by
-
Down-regulation of colon mucin production induced by Eimeria pragensis infection in mice.Front Parasitol. 2025 Jun 24;4:1621486. doi: 10.3389/fpara.2025.1621486. eCollection 2025. Front Parasitol. 2025. PMID: 40630939 Free PMC article.
-
Immunity to Cryptosporidium: insights into principles of enteric responses to infection.Nat Rev Immunol. 2024 Feb;24(2):142-155. doi: 10.1038/s41577-023-00932-3. Epub 2023 Sep 11. Nat Rev Immunol. 2024. PMID: 37697084 Free PMC article. Review.
-
Cryptosporidium uses CSpV1 to activate host type I interferon and attenuate antiparasitic defenses.Nat Commun. 2023 Mar 16;14(1):1456. doi: 10.1038/s41467-023-37129-0. Nat Commun. 2023. PMID: 36928642 Free PMC article.
-
Investigating Cryptosporidium spp. Using Genomic, Proteomic and Transcriptomic Techniques: Current Progress and Future Directions.Int J Mol Sci. 2023 Aug 16;24(16):12867. doi: 10.3390/ijms241612867. Int J Mol Sci. 2023. PMID: 37629046 Free PMC article. Review.
-
The intersection of host in vivo metabolism and immune responses to infection with kinetoplastid and apicomplexan parasites.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0016422. doi: 10.1128/mmbr.00164-22. Epub 2024 Feb 1. Microbiol Mol Biol Rev. 2024. PMID: 38299836 Free PMC article. Review.
References
-
- Osman M, Benamrouz S, Guyot K, Baydoun M, Frealle E, Chabe M, Gantois N, Delaire B, Goffard A, Aoun A, Jurdi N, Dabboussi F, Even G, Slomianny C, Gosset P, Hamze M, Creusy C, Viscogliosi E, Certad G. 2017. High association of Cryptosporidium spp. infection with colon adenocarcinoma in Lebanese patients. PLoS One 12:e0189422. 10.1371/journal.pone.0189422. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

