Genome-wide association analysis of anthracnose resistance in sorghum [Sorghum bicolor (L.) Moench]
- PMID: 34929013
- PMCID: PMC8687563
- DOI: 10.1371/journal.pone.0261461
Genome-wide association analysis of anthracnose resistance in sorghum [Sorghum bicolor (L.) Moench]
Abstract
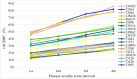
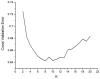
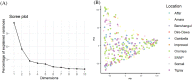
In warm-humid ago-ecologies of the world, sorghum [Sorghum bicolor (L.) Moench] production is severely affected by anthracnose disease caused by Colletotrichum sublineolum Henn. New sources of anthracnose resistance should be identified to introgress novel genes into susceptible varieties in resistance breeding programs. The objective of this study was to determine genome-wide association of Diversity Arrays Technology Sequencing (DArTseq) based single nucleotide polymorphisms (SNP) markers and anthracnose resistance genes in diverse sorghum populations for resistance breeding. Three hundred sixty-six sorghum populations were assessed for anthracnose resistance in three seasons in western Ethiopia using artificial inoculation. Data on anthracnose severity and the relative area under the disease progress curve were computed. Furthermore, the test populations were genotyped using SNP markers with DArTseq protocol. Population structure analysis and genome-wide association mapping were undertaken based on 11,643 SNPs with <10% missing data. The evaluated population was grouped into eight distinct genetic clusters. A total of eight significant (P < 0.001) marker-trait associations (MTAs) were detected, explaining 4.86-15.9% of the phenotypic variation for anthracnose resistance. Out of which the four markers were above the cutoff point. The significant MTAs in the assessed sorghum population are useful for marker-assisted selection (MAS) in anthracnose resistance breeding programs and for gene and quantitative trait loci (QTL) mapping.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
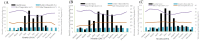

Similar articles
-
Exploring the genetic basis of anthracnose resistance in Ethiopian sorghum through a genome-wide association study.BMC Genomics. 2024 Jul 8;25(1):677. doi: 10.1186/s12864-024-10545-2. BMC Genomics. 2024. PMID: 38977981 Free PMC article.
-
Genome-Wide Association Mapping of Anthracnose (Colletotrichum sublineolum) Resistance in NPGS Ethiopian Sorghum Germplasm.G3 (Bethesda). 2019 Sep 4;9(9):2879-2885. doi: 10.1534/g3.119.400350. G3 (Bethesda). 2019. PMID: 31289022 Free PMC article.
-
Using Genotyping by Sequencing to Map Two Novel Anthracnose Resistance Loci in Sorghum bicolor.G3 (Bethesda). 2016 Jul 7;6(7):1935-46. doi: 10.1534/g3.116.030510. G3 (Bethesda). 2016. PMID: 27194807 Free PMC article.
-
Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects.Int J Mol Sci. 2018 Oct 11;19(10):3122. doi: 10.3390/ijms19103122. Int J Mol Sci. 2018. PMID: 30314374 Free PMC article. Review.
-
Elucidating Anthracnose Resistance Mechanisms in Sorghum-A Review.Phytopathology. 2020 Dec;110(12):1863-1876. doi: 10.1094/PHYTO-04-20-0132-RVW. Epub 2020 Nov 4. Phytopathology. 2020. PMID: 33100146 Review.
Cited by
-
Recent advancements in the breeding of sorghum crop: current status and future strategies for marker-assisted breeding.Front Genet. 2023 May 11;14:1150616. doi: 10.3389/fgene.2023.1150616. eCollection 2023. Front Genet. 2023. PMID: 37252661 Free PMC article. Review.
-
Sustainable Management of Major Fungal Phytopathogens in Sorghum (Sorghum bicolor L.) for Food Security: A Comprehensive Review.J Fungi (Basel). 2025 Mar 6;11(3):207. doi: 10.3390/jof11030207. J Fungi (Basel). 2025. PMID: 40137245 Free PMC article. Review.
-
Exploring the genetic basis of anthracnose resistance in Ethiopian sorghum through a genome-wide association study.BMC Genomics. 2024 Jul 8;25(1):677. doi: 10.1186/s12864-024-10545-2. BMC Genomics. 2024. PMID: 38977981 Free PMC article.
-
Transcriptome analysis provides insights into the bases of salicylic acid-induced resistance to anthracnose in sorghum.Plant Mol Biol. 2022 Sep;110(1-2):69-80. doi: 10.1007/s11103-022-01286-5. Epub 2022 Jul 6. Plant Mol Biol. 2022. PMID: 35793006
References
-
- Dahlberg J, Berenji J, Sikora V, Latkovic D. Assessing sorghum [Sorghum bicolor (L) Moench] germplasm for new traits: food, fuels and unique uses. Maydica. 2011; 56:85–92.
-
- Nida H, Girma G, Mekonen M, Lee S, Seyoum A, Dessalegn K, et al.. Identification of sorghum grain mold resistance loci through genome wide association mapping. Journal of Cereal Science. 2019; 85:295–304.
-
- Ejeta G, Gressel J. Integrating new technologies for striga control: towards ending the witch-hunt: World Scientific Publishing; 2007. 345 pp.
-
- Chala A, Brurberg MB, Tronsmo AM. Prevalence and intensity of sorghum anthracnose in Ethiopia. Journal of SAT Agricultural Research. 2007; 5:145–298.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

