Quantitative analysis of the formation of nucleoprotein complexes between HIV-1 Gag protein and genomic RNA using transmission electron microscopy
- PMID: 34929171
- PMCID: PMC8760521
- DOI: 10.1016/j.jbc.2021.101500
Quantitative analysis of the formation of nucleoprotein complexes between HIV-1 Gag protein and genomic RNA using transmission electron microscopy
Abstract
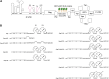
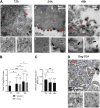
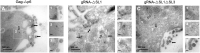
In HIV, the polyprotein precursor Gag orchestrates the formation of the viral capsid. In the current view of this viral assembly, Gag forms low-order oligomers that bind to the viral genomic RNA triggering the formation of high-ordered ribonucleoprotein complexes. However, this assembly model was established using biochemical or imaging methods that do not describe the cellular location hosting Gag-gRNA complex nor distinguish gRNA packaging in single particles. Here, we studied the intracellular localization of these complexes by electron microscopy and monitored the distances between the two partners by morphometric analysis of gold beads specifically labeling Gag and gRNA. We found that formation of these viral clusters occurred shortly after the nuclear export of the gRNA. During their transport to the plasma membrane, the distance between Gag and gRNA decreases together with an increase of gRNA packaging. Point mutations in the zinc finger patterns of the nucleocapsid domain of Gag caused an increase in the distance between Gag and gRNA as well as a sharp decrease of gRNA packaged into virions. Finally, we show that removal of stem loop 1 of the 5'-untranslated region does not interfere with gRNA packaging, whereas combined with the removal of stem loop 3 is sufficient to decrease but not abolish Gag-gRNA cluster formation and gRNA packaging. In conclusion, this morphometric analysis of Gag-gRNA cluster formation sheds new light on HIV-1 assembly that can be used to describe at nanoscale resolution other viral assembly steps involving RNA or protein-protein interactions.
Keywords: Gag; HIV-1; assembly; genomic RNA; morphometric TEM analysis; transmission electron microscopy.
Copyright © 2021. Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

