cbpManager: a web application to streamline the integration of clinical and genomic data in cBioPortal to support the Molecular Tumor Board
- PMID: 34930224
- PMCID: PMC8686377
- DOI: 10.1186/s12911-021-01719-z
cbpManager: a web application to streamline the integration of clinical and genomic data in cBioPortal to support the Molecular Tumor Board
Abstract
Background: Extensive sequencing of tumor tissues has greatly improved our understanding of cancer biology over the past years. The integration of genomic and clinical data is increasingly used to select personalized therapies in dedicated tumor boards (Molecular Tumor Boards) or to identify patients for basket studies. Genomic alterations and clinical information can be stored, integrated and visualized in the open-access resource cBioPortal for Cancer Genomics. cBioPortal can be run as a local instance enabling storage and analysis of patient data in single institutions, in the respect of data privacy. However, uploading clinical input data and genetic aberrations requires the elaboration of multiple data files and specific data formats, which makes it difficult to integrate this system into clinical practice. To solve this problem, we developed cbpManager.
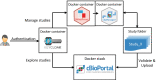
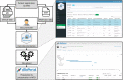
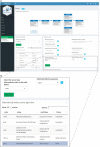
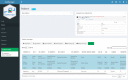
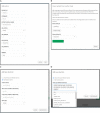
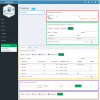
Results: cbpManager is an R package providing a web-based interactive graphical user interface intended to facilitate the maintenance of mutations data and clinical data, including patient and sample information, as well as timeline data. cbpManager enables a large spectrum of researchers and physicians, regardless of their informatics skills to intuitively create data files ready for upload in cBioPortal for Cancer Genomics on a daily basis or in batch. Due to its modular structure based on R Shiny, further data formats such as copy number and fusion data can be covered in future versions. Further, we provide cbpManager as a containerized solution, enabling a straightforward large-scale deployment in clinical systems and secure access in combination with ShinyProxy. cbpManager is freely available via the Bioconductor project at https://bioconductor.org/packages/cbpManager/ under the AGPL-3 license. It is already used at six University Hospitals in Germany (Mainz, Gießen, Lübeck, Halle, Freiburg, and Marburg).
Conclusion: In summary, our package cbpManager is currently a unique software solution in the workflow with cBioPortal for Cancer Genomics, to assist the user in the interactive generation and management of study files suited for the later upload in cBioPortal.
Keywords: Bioconductor; Clinical data; Data management; File generation; Genomic data; Molecular Tumor Board; Patient management; R; Shiny; cBioPortal.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
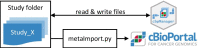

References
-
- Chang W, Cheng J, Allaire J, Sievert C, Schloerke B, Xie Y, et al. shiny: Web Application Framework for R. R package version 1.6.0. 2021. https://CRAN.R-project.org/package=shiny. Accessed 30 Jun 2021.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

