Finding genetically-supported drug targets for Parkinson's disease using Mendelian randomization of the druggable genome
- PMID: 34930919
- PMCID: PMC8688480
- DOI: 10.1038/s41467-021-26280-1
Finding genetically-supported drug targets for Parkinson's disease using Mendelian randomization of the druggable genome
Abstract
Parkinson's disease is a neurodegenerative movement disorder that currently has no disease-modifying treatment, partly owing to inefficiencies in drug target identification and validation. We use Mendelian randomization to investigate over 3,000 genes that encode druggable proteins and predict their efficacy as drug targets for Parkinson's disease. We use expression and protein quantitative trait loci to mimic exposure to medications, and we examine the causal effect on Parkinson's disease risk (in two large cohorts), age at onset and progression. We propose 23 drug-targeting mechanisms for Parkinson's disease, including four possible drug repurposing opportunities and two drugs which may increase Parkinson's disease risk. Of these, we put forward six drug targets with the strongest Mendelian randomization evidence. There is remarkably little overlap between our drug targets to reduce Parkinson's disease risk versus progression, suggesting different molecular mechanisms. Drugs with genetic support are considerably more likely to succeed in clinical trials, and we provide compelling genetic evidence and an analysis pipeline to prioritise Parkinson's disease drug development.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



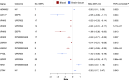
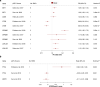
References
-
- Smietana K, Siatkowski M, Møller M. Trends in clinical success rates. Nat. Rev. Drug Discov. 2016;15:379–380. - PubMed
-
- Harrison RK. Phase II and phase III failures: 2013–2015. Nat. Rev. Drug Discov. 2016;15:817–818. - PubMed
-
- Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

