Insulin signalling in tanycytes gates hypothalamic insulin uptake and regulation of AgRP neuron activity
- PMID: 34931084
- PMCID: PMC8688146
- DOI: 10.1038/s42255-021-00499-0
Insulin signalling in tanycytes gates hypothalamic insulin uptake and regulation of AgRP neuron activity
Abstract
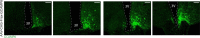
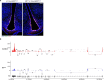
Insulin acts on neurons and glial cells to regulate systemic glucose metabolism and feeding. However, the mechanisms of insulin access in discrete brain regions are incompletely defined. Here we show that insulin receptors in tanycytes, but not in brain endothelial cells, are required to regulate insulin access to the hypothalamic arcuate nucleus. Mice lacking insulin receptors in tanycytes (IR∆Tan mice) exhibit systemic insulin resistance, while displaying normal food intake and energy expenditure. Tanycytic insulin receptors are also necessary for the orexigenic effects of ghrelin, but not for the anorexic effects of leptin. IR∆Tan mice exhibit increased agouti-related peptide (AgRP) neuronal activity, while displaying blunted AgRP neuronal adaptations to feeding-related stimuli. Lastly, a highly palatable food decreases tanycytic and arcuate nucleus insulin signalling to levels comparable to those seen in IR∆Tan mice. These changes are rooted in modifications of cellular stress responses and of mitochondrial protein quality control in tanycytes. Conclusively, we reveal a critical role of tanycyte insulin receptors in gating feeding-state-dependent regulation of AgRP neurons and systemic insulin sensitivity, and show that insulin resistance in tanycytes contributes to the pleiotropic manifestations of obesity-associated insulin resistance.
© 2022. The Author(s).
Conflict of interest statement
A.S. and T.Å.P. are current employees and stakeholders at Novo Nordisk A/S. The labelled insulin was provided by Novo Nordisk as well. The other authors declare no competing interests.
Figures

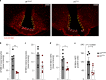





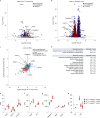

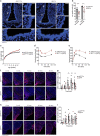
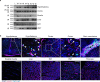
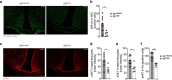

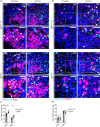
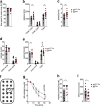
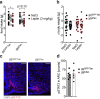
References
-
- Könner AC, Brüning JC. Selective insulin and leptin resistance in metabolic disorders. Cell Metab. 2012;16:144–152. - PubMed
-
- Brüning JC, et al. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289:2122–2125. - PubMed
-
- Könner AC, et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab. 2007;5:438–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

