Disassembly of Single Virus Capsids Monitored in Real Time with Multicycle Resistive-Pulse Sensing
- PMID: 34932317
- PMCID: PMC8784147
- DOI: 10.1021/acs.analchem.1c03855
Disassembly of Single Virus Capsids Monitored in Real Time with Multicycle Resistive-Pulse Sensing
Abstract
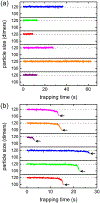
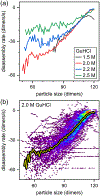
Virus assembly and disassembly are critical steps in the virus lifecycle; however, virus disassembly is much less well understood than assembly. For hepatitis B virus (HBV) capsids, disassembly of the virus capsid in the presence of guanidine hydrochloride (GuHCl) exhibits strong hysteresis that requires additional chemical energy to initiate disassembly and disrupt the capsid structure. To study disassembly of HBV capsids, we mixed T = 4 HBV capsids with 1.0-3.0 M GuHCl, monitored the reaction over time by randomly selecting particles, and measured their size with resistive-pulse sensing. Particles were cycled forward and backward multiple times to increase the observation time and likelihood of observing a disassembly event. The four-pore device used for resistive-pulse sensing produces four current pulses for each particle during translocation that improves tracking and identification of single particles and increases the precision of particle-size measurements when pulses are averaged. We studied disassembly at GuHCl concentrations below and above denaturing conditions of the dimer, the fundamental unit of HBV capsid assembly. As expected, capsids showed little disassembly at low GuHCl concentrations (e.g., 1.0 M GuHCl), whereas at higher GuHCl concentrations (≥1.5 M), capsids exhibited disassembly, sometimes as a complex series of events. In all cases, disassembly was an accelerating process, where capsids catastrophically disassembled within a few 100 ms of reaching critical stability; disassembly rates reached tens of dimers per second just before capsids fell apart. Some disassembly events exhibited metastable intermediates that appeared to lose one or more trimers of dimers in a stepwise fashion.
Figures




Similar articles
-
Characterization of Virus Capsids and Their Assembly Intermediates by Multicycle Resistive-Pulse Sensing with Four Pores in Series.Anal Chem. 2018 Jun 19;90(12):7267-7274. doi: 10.1021/acs.analchem.8b00452. Epub 2018 May 29. Anal Chem. 2018. PMID: 29708733 Free PMC article.
-
Guanidine Hydrochloride-Induced Hepatitis B Virus Capsid Disassembly Hysteresis.Biochemistry. 2024 Jun 18;63(12):1543-1552. doi: 10.1021/acs.biochem.4c00077. Epub 2024 May 24. Biochemistry. 2024. PMID: 38787909 Free PMC article.
-
Monitoring Assembly of Virus Capsids with Nanofluidic Devices.ACS Nano. 2015 Sep 22;9(9):9087-96. doi: 10.1021/acsnano.5b03231. Epub 2015 Aug 26. ACS Nano. 2015. PMID: 26266555 Free PMC article.
-
Assembly, stability and dynamics of virus capsids.Arch Biochem Biophys. 2013 Mar;531(1-2):65-79. doi: 10.1016/j.abb.2012.10.015. Epub 2012 Nov 8. Arch Biochem Biophys. 2013. PMID: 23142681 Review.
-
Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application.Viruses. 2017 Mar 21;9(3):56. doi: 10.3390/v9030056. Viruses. 2017. PMID: 28335554 Free PMC article. Review.
Cited by
-
Integrated In-Plane Nanofluidic Devices for Resistive-Pulse Sensing.Annu Rev Anal Chem (Palo Alto Calif). 2024 Jul;17(1):221-242. doi: 10.1146/annurev-anchem-061622-030223. Epub 2024 Jul 2. Annu Rev Anal Chem (Palo Alto Calif). 2024. PMID: 38608295 Free PMC article. Review.
-
pyCapsid: identifying dominant dynamics and quasi-rigid mechanical units in protein shells.Bioinformatics. 2024 Jan 2;40(1):btad761. doi: 10.1093/bioinformatics/btad761. Bioinformatics. 2024. PMID: 38113434 Free PMC article.
-
Engineering Metastability into a Virus-like Particle to Enable Triggered Dissociation.J Am Chem Soc. 2023 Feb 1;145(4):2322-2331. doi: 10.1021/jacs.2c10937. Epub 2023 Jan 18. J Am Chem Soc. 2023. PMID: 36651799 Free PMC article.
-
Solid-State Nanopore/Nanochannel Sensing of Single Entities.Top Curr Chem (Cham). 2023 Apr 27;381(4):13. doi: 10.1007/s41061-023-00425-w. Top Curr Chem (Cham). 2023. PMID: 37103594 Review.
-
Low-cost and convenient fabrication of polymer micro/nanopores with the needle punching process and their applications in nanofluidic sensing.Biomicrofluidics. 2024 Apr 1;18(2):024103. doi: 10.1063/5.0203512. eCollection 2024 Mar. Biomicrofluidics. 2024. PMID: 38571910 Free PMC article.
References
-
- Seeger C; Zoulim F; Mason WS Hepadnaviruses. In Fields Virology, Knipe DM; Howley PM, Eds. Lippincott, Williams & Wilkins: Philadelphia, 2007; Vol. 2, pp 2977–3029.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

