Rare coding variants in 35 genes associate with circulating lipid levels-A multi-ancestry analysis of 170,000 exomes
- PMID: 34932938
- PMCID: PMC8764201
- DOI: 10.1016/j.ajhg.2021.11.021
Rare coding variants in 35 genes associate with circulating lipid levels-A multi-ancestry analysis of 170,000 exomes
Abstract
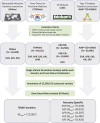
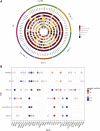
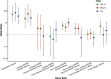
Large-scale gene sequencing studies for complex traits have the potential to identify causal genes with therapeutic implications. We performed gene-based association testing of blood lipid levels with rare (minor allele frequency < 1%) predicted damaging coding variation by using sequence data from >170,000 individuals from multiple ancestries: 97,493 European, 30,025 South Asian, 16,507 African, 16,440 Hispanic/Latino, 10,420 East Asian, and 1,182 Samoan. We identified 35 genes associated with circulating lipid levels; some of these genes have not been previously associated with lipid levels when using rare coding variation from population-based samples. We prioritize 32 genes in array-based genome-wide association study (GWAS) loci based on aggregations of rare coding variants; three (EVI5, SH2B3, and PLIN1) had no prior association of rare coding variants with lipid levels. Most of our associated genes showed evidence of association among multiple ancestries. Finally, we observed an enrichment of gene-based associations for low-density lipoprotein cholesterol drug target genes and for genes closest to GWAS index single-nucleotide polymorphisms (SNPs). Our results demonstrate that gene-based associations can be beneficial for drug target development and provide evidence that the gene closest to the array-based GWAS index SNP is often the functional gene for blood lipid levels.
Keywords: association; cholesterol; exome sequencing; gene-based association; lipid.
Copyright © 2021 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests for the present work. P.N. reports investigator-initiated grants from Amgen, Apple, and Boston Scientific; is a scientific advisor to Apple, Blackstone Life Sciences, and Novartis; and has spousal employment at Vertex, all unrelated to the present work. A.V.K. has served as a scientific advisor to Sanofi, Medicines Company, Maze Pharmaceuticals, Navitor Pharmaceuticals, Verve Therapeutics, Amgen, and Color; received speaking fees from Illumina, MedGenome, Amgen, and the Novartis Institute for Biomedical Research; received sponsored research agreements from the Novartis Institute for Biomedical Research and IBM Research; and reports a patent related to a genetic risk predictor (20190017119). C.J.W.’s spouse is employed at Regeneron. L.E.S. is currently an employee of Celgene/Bristol Myers Squibb. Celgene/Bristol Myers Squibb had no role in the funding, design, conduct, and interpretation of this study. M.E.M. receives funding from Regeneron unrelated to this work. E.E.K. has received speaker honoraria from Illumina, Inc and Regeneron Pharmaceuticals. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. L.A.C. has consulted with the Dyslipidemia Foundation on lipid projects in the Framingham Heart Study. P.T.E. is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular disease. P.T.E. has consulted for Bayer AG, Novartis, MyoKardia, and Quest Diagnostics. S.A.L. receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer AG, Boehringer Ingelheim, Fitbit, and IBM and has consulted for Bristol Myers Squibb/Pfizer, Bayer AG, and Blackstone Life Sciences. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. M.I.M. has served on advisory panels for Pfizer, NovoNordisk, and Zoe Global and has received honoraria from Merck, Pfizer, Novo Nordisk, and Eli Lilly and research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier, and Takeda. As of June 2019, M.I.M. is an employee of Genentech and a holder of Roche stock. M.E.J. holds shares in Novo Nordisk A/S. H.M.K. is an employee of Regeneron Pharmaceuticals; he owns stock and stock options for Regeneron Pharmaceuticals. M.E.J. has received research grants form Astra Zeneca, Boehringer Ingelheim, Amgen, and Sanofi. S.K. is founder of Verve Therapeutics.
Figures
References
-
- Peloso G.M., Auer P.L., Bis J.C., Voorman A., Morrison A.C., Stitziel N.O., Brody J.A., Khetarpal S.A., Crosby J.R., Fornage M., et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 2014;94:223–232. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK093757/DK/NIDDK NIH HHS/United States
- RG/13/13/30194/BHF_/British Heart Foundation/United Kingdom
- R01 HL109946/HL/NHLBI NIH HHS/United States
- R03 DK131249/DK/NIDDK NIH HHS/United States
- K23 DK114551/DK/NIDDK NIH HHS/United States
- CS/14/2/30841/BHF_/British Heart Foundation/United Kingdom
- U01 DK062370/DK/NIDDK NIH HHS/United States
- UM1 DK078616/DK/NIDDK NIH HHS/United States
- RG/18/10/33842/BHF_/British Heart Foundation/United Kingdom
- R01 DK125490/DK/NIDDK NIH HHS/United States
- R01 HL113323/HL/NHLBI NIH HHS/United States
- R01 DK127636/DK/NIDDK NIH HHS/United States
- RG/18/13/33946/BHF_/British Heart Foundation/United Kingdom
- R01 AG058921/AG/NIA NIH HHS/United States
- R01 HL153805/HL/NHLBI NIH HHS/United States
- R01 DK072193/DK/NIDDK NIH HHS/United States
- P30 DK079626/DK/NIDDK NIH HHS/United States
- R01 HL127564/HL/NHLBI NIH HHS/United States
- R01 DK062370/DK/NIDDK NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 HL142711/HL/NHLBI NIH HHS/United States
- R35 HL135818/HL/NHLBI NIH HHS/United States
- CH/12/2/29428/BHF_/British Heart Foundation/United Kingdom
- P01 HL045522/HL/NHLBI NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- R03 HL154284/HL/NHLBI NIH HHS/United States
- K08 HG010155/HG/NHGRI NIH HHS/United States
- K01 HL135405/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

