Targeting uPA-uPAR interaction to improve intestinal epithelial barrier integrity in inflammatory bowel disease
- PMID: 34933179
- PMCID: PMC8688562
- DOI: 10.1016/j.ebiom.2021.103758
Targeting uPA-uPAR interaction to improve intestinal epithelial barrier integrity in inflammatory bowel disease
Abstract
Background: Loss of intestinal epithelial barrier integrity is a critical component of Inflammatory Bowel Disease (IBD) pathogenesis. Co-expression regulation of ligand-receptor pairs in IBD mucosa has not been systematically studied. Targeting ligand-receptor pairs which are induced in IBD mucosa and function in intestinal epithelial barrier integrity may provide novel therapeutics for IBD.
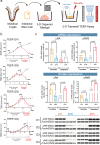
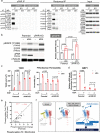
Methods: We performed transcriptomic meta-analysis on public IBD datasets combined with cell surface protein-protein-interaction (PPI) databases. We explored primary human/mouse intestinal organoids and Caco-2 cells for expression and function studies of uPA-uPAR (prime hits from the meta-analysis). Epithelial barrier integrity was measured by Trans-Epithelial Electrical Resistance (TEER), FITC-Dextran permeability and tight junction assessment. Genetic (CRISPR, siRNA and KO mice) and pharmacological (small molecules, neutralizing antibody and peptide inhibitors) approaches were applied. Mice deficient of uPAR were studied using the Dextran Sulfate Sodium (DSS)-induced colitis model.
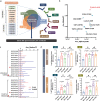
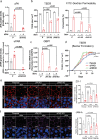
Findings: The IBD ligand-receptor meta-analysis led to the discovery of a coordinated upregulation of uPA and uPAR in IBD mucosa. Both genes were significantly upregulated during epithelial barrier breakdown in primary intestinal organoids and decreased during barrier formation. Genetic inhibition of uPAR or uPA, or pharmacologically blocking uPA-uPAR interaction protects against cytokine-induced barrier breakdown. Deficiency of uPAR in epithelial cells leads to enhanced EGF/EGFR signalling, a known regulator of epithelial homeostasis and repair. Mice deficient of uPAR display improved intestinal barrier function in vitro and during DSS-induced colitis in vivo.
Interpretation: Our findings suggest that blocking uPA-uPAR interaction via pharmacological agents protects the epithelial barrier from inflammation-induced damage, indicating a potential therapeutic target for IBD.
Funding: The study was funded by Boehringer Ingelheim.
Keywords: IBD; Intestinal epithelial barrier; Organoid; uPA; uPAR.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests During the study, all the authors were Boehringer Ingelheim employees. No author has any further financial, professional, or personal disclosures.
Figures


References
-
- McGuckin MA, Eri R, Simms LA, Florin TH, Radford-Smith G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15(1):100–113. - PubMed
-
- Neurath MF, Travis SP. Mucosal healing in inflammatory bowel diseases: a systematic review. Gut. 2012;61(11):1619–1635. - PubMed
-
- Neurath MF. New targets for mucosal healing and therapy in inflammatory bowel diseases. Mucosal Immunol. 2014;7(1):6–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

