Early Tumor-Immune Microenvironmental Remodeling and Response to First-Line Fluoropyrimidine and Platinum Chemotherapy in Advanced Gastric Cancer
- PMID: 34933901
- PMCID: PMC9387589
- DOI: 10.1158/2159-8290.CD-21-0888
Early Tumor-Immune Microenvironmental Remodeling and Response to First-Line Fluoropyrimidine and Platinum Chemotherapy in Advanced Gastric Cancer
Abstract
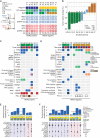
Chemotherapy is ubiquitous in first-line treatment of advanced gastric cancer, yet responses are heterogeneous, and little is known about mediators of chemotherapy response. To move forward, an understanding of the effects of standard chemotherapy on the tumor-immune microenvironment (TME) is needed. Coupling whole-exome sequencing, bulk RNA and single-cell transcriptomics from paired pretreatment and on-treatment samples in treatment-naïve patients with HER2-positive and HER2-negative gastric cancer, we define features associated with response to platinum-based chemotherapy. Response was associated with on-treatment TME remodeling including natural killer (NK) cell recruitment, decreased tumor-associated macrophages, M1-macrophage repolarization, and increased effector T-cell infiltration. Among chemotherapy nonresponders, we observed low/absent PD-L1 expression or modulation, on-treatment increases in Wnt signaling, B-cell infiltration, and LAG3-expressing T cells coupled to an exodus of dendritic cells. We did not observe significant genomic changes in early on-treatment sampling. We provide a map of on-treatment TME modulation with standard chemotherapy and nominate candidate future approaches.
Significance: Using paired pretreatment and on-treatment samples during standard first-line chemotherapy, we identify chemotherapy-induced NK-cell infiltration, macrophage repolarization, and increased antigen presentation among responders. Increased LAG3 expression and decreased dendritic cell abundance were seen in nonresponders, emphasizing remodeling of the TME during chemotherapy response and resistance. This article is highlighted in the In This Issue feature, p. 873.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures





Comment in
- Cancer Discov. 12:873.
- Cancer Discov. 12:873.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. - PubMed
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo Met al. . Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. - PubMed
-
- Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki Aet al. . Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010;376:687–97. - PubMed
-
- Cunningham D, Starling N, Rao S, Iveson T, Nicolson M, Coxon Fet al. . Capecitabine and oxaliplatin for advanced esophagogastric cancer. N Engl J Med 2008;358:36–46. - PubMed
-
- Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi Met al. . S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): A phase III trial. Lancet Oncol 2008;9:215–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

