Tetramerization of STAT5 promotes autoimmune-mediated neuroinflammation
- PMID: 34934004
- PMCID: PMC8719886
- DOI: 10.1073/pnas.2116256118
Tetramerization of STAT5 promotes autoimmune-mediated neuroinflammation
Abstract
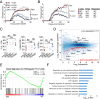
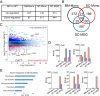
Signal tranducer and activator of transcription 5 (STAT5) plays a critical role in mediating cellular responses following cytokine stimulation. STAT proteins critically signal via the formation of dimers, but additionally, STAT tetramers serve key biological roles, and we previously reported their importance in T and natural killer (NK) cell biology. However, the role of STAT5 tetramerization in autoimmune-mediated neuroinflammation has not been investigated. Using the STAT5 tetramer-deficient Stat5a-Stat5b N-domain double knockin (DKI) mouse strain, we report here that STAT5 tetramers promote the pathogenesis of experimental autoimmune encephalomyelitis (EAE). The mild EAE phenotype observed in DKI mice correlates with the impaired extravasation of pathogenic T-helper 17 (Th17) cells and interactions between Th17 cells and monocyte-derived cells (MDCs) in the meninges. We further demonstrate that granulocyte-macrophage colony-stimulating factor (GM-CSF)-mediated STAT5 tetramerization regulates the production of CCL17 by MDCs. Importantly, CCL17 can partially restore the pathogenicity of DKI Th17 cells, and this is dependent on the activity of the integrin VLA-4. Thus, our study reveals a GM-CSF-STAT5 tetramer-CCL17 pathway in MDCs that promotes autoimmune neuroinflammation.
Keywords: CCL17; STAT5 tetramers; Th17; autoimmunity; monocytes.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Compston A., Coles A., Multiple sclerosis. Lancet 372, 1502–1517 (2008). - PubMed
-
- Dendrou C. A., Fugger L., Friese M. A., Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 15, 545–558 (2015). - PubMed
-
- Mundt S., et al. , Conventional DCs sample and present myelin antigens in the healthy CNS and allow parenchymal T cell entry to initiate neuroinflammation. Sci. Immunol. 4, eaau8380 (2019). - PubMed
-
- Prinz M., Priller J., The role of peripheral immune cells in the CNS in steady state and disease. Nat. Neurosci. 20, 136–144 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

