Involvement of the zebrafish trrap gene in craniofacial development
- PMID: 34934055
- PMCID: PMC8692476
- DOI: 10.1038/s41598-021-03123-z
Involvement of the zebrafish trrap gene in craniofacial development
Abstract
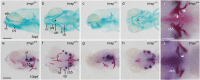
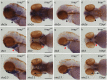
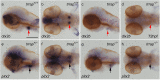
Trrap (transformation/transcription domain-associated protein) is a component shared by several histone acetyltransferase (HAT) complexes and participates in transcriptional regulation and DNA repair; however, the developmental functions of Trrap in vertebrates are not fully understood. Recently, it has been reported that human patients with genetic mutations in the TRRAP gene show various symptoms, including facial dysmorphisms, microcephaly and global developmental delay. To investigate the physiological functions of Trrap, we established trrap gene-knockout zebrafish and examined loss-of-function phenotypes in the mutants. The trrap zebrafish mutants exhibited smaller eyes and heads than the wild-type zebrafish. The size of the ventral pharyngeal arches was reduced and the mineralization of teeth was impaired in the trrap mutants. Whole-mount in situ hybridization analysis revealed that dlx3 expression was narrowly restricted in the developing ventral pharyngeal arches, while dlx2b expression was diminished in the trrap mutants. These results suggest that trrap zebrafish mutants are useful model organisms for a human disorder associated with genetic mutations in the human TRRAP gene.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


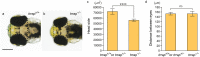
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

