Mandipropamid as a chemical inducer of proximity for in vivo applications
- PMID: 34934192
- PMCID: PMC8709788
- DOI: 10.1038/s41589-021-00922-3
Mandipropamid as a chemical inducer of proximity for in vivo applications
Abstract
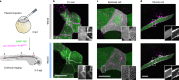
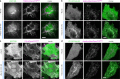
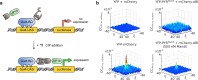
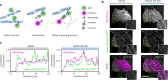
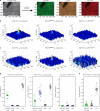
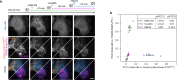
Direct control of protein interactions by chemically induced protein proximity holds great potential for both cell and synthetic biology as well as therapeutic applications. Low toxicity, orthogonality and excellent cell permeability are important criteria for chemical inducers of proximity (CIPs), in particular for in vivo applications. Here, we present the use of the agrochemical mandipropamid (Mandi) as a highly efficient CIP in cell culture systems and living organisms. Mandi specifically induces complex formation between a sixfold mutant of the plant hormone receptor pyrabactin resistance 1 (PYR1) and abscisic acid insensitive (ABI). It is orthogonal to other plant hormone-based CIPs and rapamycin-based CIP systems. We demonstrate the applicability of the Mandi system for rapid and efficient protein translocation in mammalian cells and zebrafish embryos, protein network shuttling and manipulation of endogenous proteins.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Spencer D, Wandless T, Schreiber S, Crabtree G. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. - PubMed
-
- Pruschy MN, et al. Mechanistic studies of a signaling pathway activated by the organic dimerizer FK1012. Chem. Biol. 1994;1:163–172. - PubMed
-
- Klemm JD, Beals CR, Crabtree GR. Rapid targeting of nuclear proteins to the cytoplasm. Curr. Biol. 1997;7:638–644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

