Host cell entry mediators implicated in the cellular tropism of SARS‑CoV‑2, the pathophysiology of COVID‑19 and the identification of microRNAs that can modulate the expression of these mediators (Review)
- PMID: 34935057
- PMCID: PMC8722767
- DOI: 10.3892/ijmm.2021.5075
Host cell entry mediators implicated in the cellular tropism of SARS‑CoV‑2, the pathophysiology of COVID‑19 and the identification of microRNAs that can modulate the expression of these mediators (Review)
Abstract
The pathophysiology of coronavirus disease 2019 (COVID‑19) is mainly dependent on the underlying mechanisms that mediate the entry of severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) into the host cells of the various human tissues/organs. Recent studies have indicated a higher order of complexity of the mechanisms of infectivity, given that there is a wide‑repertoire of possible cell entry mediators that appear to co‑localise in a cell‑ and tissue‑specific manner. The present study provides an overview of the 'canonical' SARS‑CoV‑2 mediators, namely angiotensin converting enzyme 2, transmembrane protease serine 2 and 4, and neuropilin‑1, expanding on the involvement of novel candidates, including glucose‑regulated protein 78, basigin, kidney injury molecule‑1, metabotropic glutamate receptor subtype 2, ADAM metallopeptidase domain 17 (also termed tumour necrosis factor‑α convertase) and Toll‑like receptor 4. Furthermore, emerging data indicate that changes in microRNA (miRNA/miR) expression levels in patients with COVID‑19 are suggestive of further complexity in the regulation of these viral mediators. An in silico analysis revealed 160 candidate miRNAs with potential strong binding capacity in the aforementioned genes. Future studies should concentrate on elucidating the association between the cellular tropism of the SARS‑CoV‑2 cell entry mediators and the mechanisms through which they might affect the clinical outcome. Finally, the clinical utility as a biomarker or therapeutic target of miRNAs in the context of COVID‑19 warrants further investigation.
Keywords: ACE2; ADAM17; COVID‑19; GRP78; NPR1; SARS‑CoV2; TLR4; TMPRSS2; TMPRSS4; miRNAs.
Conflict of interest statement
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that they have no competing interests.
Figures

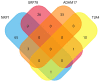
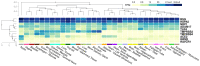
References
-
- Osuchowski MF, Winkler MS, Skirecki T, Cajander S, Shankar-Hari M, Lachmann G, Monneret G, Venet F, Bauer M, Brunkhorst FM, et al. The COVID-19 puzzle: Deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med. 2021;9:622–642. doi: 10.1016/S2213-2600(21)00218-6. - DOI - PMC - PubMed
-
- Murgolo N, Therien AG, Howell B, Klein D, Koeplinger K, Lieberman LA, Adam GC, Flynn J, McKenna P, Swaminathan G, et al. SARS-CoV-2 tropism, entry, replication, and propagation: Considerations for drug discovery and development. PLoS Pathog. 2021;17:e1009225. doi: 10.1371/journal.ppat.1009225. - DOI - PMC - PubMed
-
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

