Helical ordering of envelope-associated proteins and glycoproteins in respiratory syncytial virus
- PMID: 34935163
- PMCID: PMC8804925
- DOI: 10.15252/embj.2021109728
Helical ordering of envelope-associated proteins and glycoproteins in respiratory syncytial virus
Abstract
Human respiratory syncytial virus (RSV) causes severe respiratory illness in children and the elderly. Here, using cryogenic electron microscopy and tomography combined with computational image analysis and three-dimensional reconstruction, we show that there is extensive helical ordering of the envelope-associated proteins and glycoproteins of RSV filamentous virions. We calculated a 16 Å resolution sub-tomogram average of the matrix protein (M) layer that forms an endoskeleton below the viral envelope. These data define a helical lattice of M-dimers, showing how M is oriented relative to the viral envelope. Glycoproteins that stud the viral envelope were also found to be helically ordered, a property that was coordinated by the M-layer. Furthermore, envelope glycoproteins clustered in pairs, a feature that may have implications for the conformation of fusion (F) glycoprotein epitopes that are the principal target for vaccine and monoclonal antibody development. We also report the presence, in authentic virus infections, of N-RNA rings packaged within RSV virions. These data provide molecular insight into the organisation of the virion and the mechanism of its assembly.
Keywords: cryo-EM; cryo-ET; glycoprotein; matrix protein; virus structure.
© 2021 The Authors Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Immunofluorescence imaging of vero cells infected with RSV at 72 h post‐infection shows an abundance of filamentous particles. Virus particles were identified by labelling with an antibody that targeted the nucleocapsid protein (green). Phalloidin was used to localise actin (red), and DAPI reagent was used to stain cell nuclei (blue).
Cryo‐EM image of a short section of RSV filament.
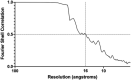
The power‐spectrum for the filament shown in (B) is shown, Fourier‐Bessel analysis was used to identify a putative helical lattice, the positions of principal maxima are indicated by boxes, and however, one was found to be missing (green box). The lattice was only able to be identified using higher‐order reflections.
Masking and Fourier synthesis led to the reconstitution of a one‐sided, filtered image, showing the presence of helical tracks and strong density lining the inner leaflet of the viral envelope (inset).

- A
Propagating RSV directly on the TEM grid led to improved preservation of filamentous morphology.
- B, C
Despite the gentle preparation methods, many virions showed signs of disruption such as varicosities. The areas indicated by purple boxes were imaged to collect tilt‐series. The filament in panel (A) is shown in Fig 3A, the filament in panel (B) is shown in Fig 2G, and the filament shown in panel (C) is shown in Fig 3B.
- D, E
(D) A central slice through a denoised tomogram shows a complex viral envelope, shown at higher magnification in (E). Glycoprotein spikes are densely packed, giving a picket‐fence like appearance when viewed from the side (highlighted in mauve). The lipid bilayer is highlighted in pale blue, the matrix layer in orange, and the less well‐ordered protein layer previously hypothesised to consist of M2‐1 is shaded in dark blue.
- F
Tilt‐series were collected targeting straight regions of filamentous virions; however, some tomograms captured the virion ends, which were found to be hemispherical but rather flattened. In some cases (as shown here), the matrix layer was intact and contiguous at the filament ends (blue arrows), while some virions showed an absence of M.
- G
Loss of virion integrity was accompanied by discontinuities in the matrix layer (green arrows).

Central slices through tomograms showed that virions contained helical nucleocapsids having the characteristic herringbone morphology (blue arrow).
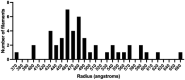
Many virions also contain large numbers of ring‐shaped assemblies.
A close‐up view of these structures reveals the presence of radial spokes (pink arrows). These structures are morphologically very similar to previously described decameric and undecameric rings produced by recombinant expression of RSV N.

- A, B
(A) A slice through the glycoprotein spikes of a filamentous RSV virion shows that they are helically ordered. Long‐range ordering of the glycoprotein spikes is evident, as is clustering of spikes in pairs (highlighted in (B)).
- C–F
In virions that are sparsely decorated with glycoprotein spikes, clustering in pairs is even more apparent.
- G
A central section through a tomogram of a pleomorphic RSV virion showing the presence of both herringbone nucleocapsids and N‐RNA rings (blue arrows), flattened, thicker membranes that likely have a matrix layer (green arrow), and tightly packed glycoprotein spikes (pink arrows).
- H, I
A section through the envelope region of the same particle shows the presence of a honeycomb‐like array of densities that may be an alternate arrangement of glycoprotein spikes, and discrete densities are seen (purple arrow) that are likely individual glycoprotein spikes.

- A, B
Tomograms of a pleomorphic particle show extensive patches of glycoprotein arrays with hexagonal packing. Slices are shown at different z values through the same particle.

Viewed from the filament interior, a regular array of density is seen in the envelope layer attributed to the matrix protein.
The average is tilted 45° towards the viewer.
The average is tilted a further 45°, giving a view of the lipid bilayer. The inner leaflet of the bilayer is only weakly resolved compared to the outer leaflet.
Viewed from the virion exterior at a lower isosurface threshold, stripes of weak, noisy density are seen. This is likely incoherent averaging of glycoprotein spike density.
Tilting the reconstruction towards the viewer by 45° shows that the inner leaflet contacts the M‐layer.
A fourth density layer is now visible, under the matrix. This density has been previously attributed to the RNA polymerase co‐factor M2‐1. Like the glycoprotein density at the exterior of the envelope, the M2‐1 density is weak and incoherently averaged.

- A, B
The published X‐ray structure for RSV M shows that it forms a dimer (PDB accession number 4V23).
- C
In the X‐ray structure, M‐dimers pack as stacked sheets in the (010) plane of the C2 crystals.
- D
Docking the M‐dimer into the 3D reconstruction shows a curved lattice with similar packing in virio to the planar sheets seen in the crystal structure.
- E
Close‐up view of the docked M coordinates.
- F
Measurements of the lattice spacings of M‐dimers in the docked model of the matrix layer. A low‐pitch helix corresponding to the (1,1) direction of the planar lattice (shown in panel C) has a spacing of 82 Å. Helix strands corresponding to the a and c axes in the C2 unit cell of the X‐ray structure have spacings of 54 Å and 66 Å respectively.

- A
A section through the sub‐tomogram average reconstruction with fitted coordinates for M shows the orientation of the M‐dimer relative to the inner leaflet of the lipid bilayer.
- B–E
(B and C) A solvent excluded surface representation of the matrix layer is presented coloured to show the electrostatic potential. It shows that the surface facing the lipid bilayer is positively charged, while the surface facing the virion interior presents stripes of negative charge (D and E).
- F
A view of the docked model placed within a transparent isosurface of the sub‐tomogram average reconstruction and viewed at an isosurface threshold of 0 from the virion exterior shows that the noisy weak density that we attribute to the viral glycoprotein spikes aligns with the (1,1) helix (alternate strands are coloured pink and blue).
References
-
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW (2006) Crystal structure of the rabies virus nucleoprotein‐RNA complex. Science 313: 360–363 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U105178939/MRC_/Medical Research Council/United Kingdom
- 102463/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- MR/M000451/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/7/MRC_/Medical Research Council/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC-A025-5PL41/MRC_/Medical Research Council/United Kingdom
- 099786/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- R01 AI113321/AI/NIAID NIH HHS/United States
- MC_U105184322/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/9/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

