Alterations of human skin microbiome and expansion of antimicrobial resistance after systemic antibiotics
- PMID: 34936382
- PMCID: PMC8878148
- DOI: 10.1126/scitranslmed.abd8077
Alterations of human skin microbiome and expansion of antimicrobial resistance after systemic antibiotics
Abstract
Although systemic antibiotics are critical in controlling infections and reducing morbidity and mortality, overuse of antibiotics is presumed to contribute to negative repercussions such as selection of antimicrobial-resistant organisms and collateral damage to commensal microbes. In a prospective, randomized study of four clinically relevant antibiotic regimens [doxycycline (20 mg or 100 mg), cephalexin, or trimethoprim/sulfamethoxazole], we investigated microbial alterations on skin after administration of systemic antibiotics to healthy human volunteers. Samples from different skin and oral sites, as well as stool, were collected before, during, and up to 1 year after antibiotic use, and shotgun metagenomic sequencing was performed. Taxonomic analysis showed that subjects receiving doxycycline 100 mg and trimethoprim/sulfamethoxazole (TMP/SMX) exhibited greater changes to their skin microbial communities, as compared to those receiving other regimens or untreated controls. Oral and stool microbiota also demonstrated fluctuations after antibiotics. Bacterial culturing in combination with whole-genome sequencing revealed specific emergence, expansion, and persistence of antibiotic-resistant staphylococci harboring tetK or tetL and dfrC or dfrG genes in all subjects who received doxycycline 100 mg or TMP/SMX, respectively. Last, analysis of metagenomic data revealed an increase of genes involved in gene mobilization, indicating stress responses of microbes to antibiotics. Collectively, these findings demonstrate direct, long-lasting effects of antibiotics on skin microbial communities, highlighting the skin microbiome as a site for the development and persistence of antibiotic resistance and the risks of overprescribing.
Conflict of interest statement
Figures

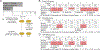


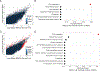
References
-
- Centers for Disease Control and Prevention. Outpatient antibiotic prescriptions — United States. (2017).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

