ATAD3A has a scaffolding role regulating mitochondria inner membrane structure and protein assembly
- PMID: 34936866
- PMCID: PMC8785211
- DOI: 10.1016/j.celrep.2021.110139
ATAD3A has a scaffolding role regulating mitochondria inner membrane structure and protein assembly
Abstract
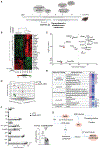
The ATPase Family AAA Domain Containing 3A (ATAD3A), is a mitochondrial inner membrane protein conserved in metazoans. ATAD3A has been associated with several mitochondrial functions, including nucleoid organization, cholesterol metabolism, and mitochondrial translation. To address its primary role, we generated a neuronal-specific conditional knockout (Atad3 nKO) mouse model, which developed a severe encephalopathy by 5 months of age. Pre-symptomatic mice showed aberrant mitochondrial cristae morphogenesis in the cortex as early as 2 months. Using a multi-omics approach in the CNS of 2-to-3-month-old mice, we found early alterations in the organelle membrane structure. We also show that human ATAD3A associates with different components of the inner membrane, including OXPHOS complex I, Letm1, and prohibitin complexes. Stochastic Optical Reconstruction Microscopy (STORM) shows that ATAD3A is regularly distributed along the inner mitochondrial membrane, suggesting a critical structural role in inner mitochondrial membrane and its organization, most likely in an ATPase-dependent manner.
Keywords: ATAD3; cardiolipin; cristae; inner membrane; mitochondria.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflict of interests.
Figures
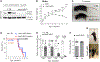




References
-
- Antonicka H, Lin ZY, Janer A, Aaltonen MJ, Weraarpachai W, Gingras AC, and Shoubridge EA (2020). A High-Density Human Mitochondrial Proximity Interaction Network. Cell Metab. 32, 479–497.e9. - PubMed
-
- Austin S, and Nowikovsky K (2019). LETM1: Essential for Mitochondrial Biology and Cation Homeostasis? Trends Biochem. Sci 44, 648–658. - PubMed
-
- Baudier J (2018). ATAD3 proteins: brokers of a mitochondria-endoplasmic reticulum connection in mammalian cells. Biol. Rev. Camb. Philos. Soc 93, 827–844. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

