Human embryonic genome activation initiates at the one-cell stage
- PMID: 34936886
- PMCID: PMC8826644
- DOI: 10.1016/j.stem.2021.11.012
Human embryonic genome activation initiates at the one-cell stage
Abstract
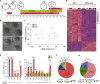
In human embryos, the initiation of transcription (embryonic genome activation [EGA]) occurs by the eight-cell stage, but its exact timing and profile are unclear. To address this, we profiled gene expression at depth in human metaphase II oocytes and bipronuclear (2PN) one-cell embryos. High-resolution single-cell RNA sequencing revealed previously inaccessible oocyte-to-embryo gene expression changes. This confirmed transcript depletion following fertilization (maternal RNA degradation) but also uncovered low-magnitude upregulation of hundreds of spliced transcripts. Gene expression analysis predicted embryonic processes including cell-cycle progression and chromosome maintenance as well as transcriptional activators that included cancer-associated gene regulators. Transcription was disrupted in abnormal monopronuclear (1PN) and tripronuclear (3PN) one-cell embryos. These findings indicate that human embryonic transcription initiates at the one-cell stage, sooner than previously thought. The pattern of gene upregulation promises to illuminate processes involved at the onset of human development, with implications for epigenetic inheritance, stem-cell-derived embryos, and cancer.
Keywords: embryonic genome activation (EGA); fertilization; human one-cell embryo; single-cell RNA-seq; totipotency; transcriptome; zygote.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Alizadeh Z., Kageyama S.-I., Aoki F. Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol. Reprod. Dev. 2005;72:281–290. - PubMed
-
- Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum. Reprod. 2011;26:1270–1283. - PubMed
-
- Amanai M., Brahmajosyula M., Perry A.C.F. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol. Reprod. 2006;75:877–884. - PubMed
-
- Asami M., Lam B.Y.H., Hoffmann M., Suzuki T., Lu X., VerMilyea M.D., Yoshida N., Ma M.K., Rainbow K., Braun S., et al. Mouse fertilization triggers a conserved transcription program in one-cell embryos. bioRxiv. 2020 doi: 10.1101/2020.09.15.298018. - DOI
Publication types
MeSH terms
Grants and funding
- BB/S017593/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UU_00014/5/MRC_/Medical Research Council/United Kingdom
- MC_UU_12012/5/MRC_/Medical Research Council/United Kingdom
- G1000839/MRC_/Medical Research Council/United Kingdom
- MR/S026193/1/MRC_/Medical Research Council/United Kingdom
- MR/N020294/1/MRC_/Medical Research Council/United Kingdom
- MR/N000080/1/MRC_/Medical Research Council/United Kingdom
- BB/P009506/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UU_00014/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 208363/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

