Enhanced fitness of SARS-CoV-2 variant of concern Alpha but not Beta
- PMID: 34937050
- PMCID: PMC8828469
- DOI: 10.1038/s41586-021-04342-0
Enhanced fitness of SARS-CoV-2 variant of concern Alpha but not Beta
Abstract
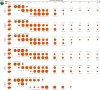
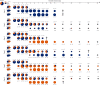
Emerging variants of concern (VOCs) are driving the COVID-19 pandemic1,2. Experimental assessments of replication and transmission of major VOCs and progenitors are needed to understand the mechanisms of replication and transmission of VOCs3. Here we show that the spike protein (S) from Alpha (also known as B.1.1.7) and Beta (B.1.351) VOCs had a greater affinity towards the human angiotensin-converting enzyme 2 (ACE2) receptor than that of the progenitor variant S(D614G) in vitro. Progenitor variant virus expressing S(D614G) (wt-S614G) and the Alpha variant showed similar replication kinetics in human nasal airway epithelial cultures, whereas the Beta variant was outcompeted by both. In vivo, competition experiments showed a clear fitness advantage of Alpha over wt-S614G in ferrets and two mouse models-the substitutions in S were major drivers of the fitness advantage. In hamsters, which support high viral replication levels, Alpha and wt-S614G showed similar fitness. By contrast, Beta was outcompeted by Alpha and wt-S614G in hamsters and in mice expressing human ACE2. Our study highlights the importance of using multiple models to characterize fitness of VOCs and demonstrates that Alpha is adapted for replication in the upper respiratory tract and shows enhanced transmission in vivo in restrictive models, whereas Beta does not overcome Alpha or wt-S614G in naive animals.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

