Evaluation of Three Quantitative Anti-SARS-CoV-2 Antibody Immunoassays
- PMID: 34937195
- PMCID: PMC8694121
- DOI: 10.1128/spectrum.01376-21
Evaluation of Three Quantitative Anti-SARS-CoV-2 Antibody Immunoassays
Abstract
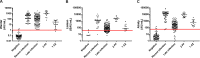
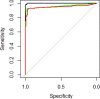
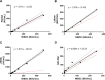
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 and caused a dramatic pandemic. Serological assays are used to check for immunization and assess herd immunity. We evaluated commercially available assays designed to quantify antibodies directed to the SARS-CoV-2 Spike (S) antigen, either total (Wantaï SARS-CoV-2 Ab ELISA) or IgG (SARS-CoV-2 IgG II Quant on Alinity, Abbott, and Liaison SARS-CoV-2 TrimericS IgG, Diasorin). The specificities of the Wantaï, Alinity, and Liaison assays were evaluated using 100 prepandemic sera and were 98, 99, and 97%, respectively. The sensitivities of all three were around 100% when tested on 35 samples taken 15 to 35 days postinfection. They were less sensitive for 150 sera from late infections (>180 days). Using the first WHO international standard (NIBSC), we showed that the Wantai results were concordant with the NIBSC values, while Liaison and Alinity showed a proportional bias of 1.3 and 7, respectively. The results of the 3 immunoassays were significantly globally pairwise correlated and for late infection sera (P < 0.001). They were correlated for recent infection sera measured with Alinity and Liaison (P < 0.001). However, the Wantai results of recent infections were not correlated with those from Alinity or Liaison. All the immunoassay results were significantly correlated with the neutralizing antibody titers obtained using a live virus neutralization assay with the B1.160 SARS-CoV-2 strain. These assays will be useful once the protective anti-SARS-CoV-2 antibody titer has been determined. IMPORTANCE Standardization and correlation with virus neutralization assays are critical points to compare the performance of serological assays designed to quantify anti-SARS-CoV-2 antibodies in order to identify their optimal use. We have evaluated three serological immunoassays based on the virus spike antigen that detect anti-SARS-CoV-2 antibodies: a microplate assay and two chemiluminescent assays performed with Alinity (Abbott) and Liaison (Diasorin) analysers. We used an in-house live virus neutralization assay and the first WHO international standard to assess the comparison. This study could be useful to determine guidelines on the use of serological results to manage vaccination and treatment with convalescent plasma or monoclonal antibodies.
Keywords: COVID; SARS-CoV-2; binding antibodies; immunoassay; neutralizing antibodies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Olbrich L, Castelletti N, Schälte Y, Garí M, Pütz P, Bakuli A, Pritsch M, Kroidl I, Saathoff E, Guggenbuehl Noller JM, Fingerle V, Le Gleut R, Gilberg L, Brand I, Falk P, Markgraf A, Deák F, Riess F, Diefenbach M, Eser T, Weinauer F, Martin S, Quenzel E-M, Becker M, Durner J, Girl P, Müller K, Radon K, Fuchs C, Wölfel R, Hasenauer J, Hoelscher M, Wieser A, On Behalf Of The KoCo-Study Group null. 2021. Head-to-head evaluation of seven different seroassays including direct viral neutralisation in a representative cohort for SARS-CoV-2. J Gen Virol 102. doi:10.1099/jgv.0.001653. - DOI - PMC - PubMed
-
- Montesinos I, Dahma H, Wolff F, Dauby N, Delaunoy S, Wuyts M, Detemmerman C, Duterme C, Vandenberg O, Martin C, Hallin M. 2021. Neutralizing antibody responses following natural SARS-CoV-2 infection: Dynamics and correlation with commercial serologic tests. J Clin Virol 144:104988. doi:10.1016/j.jcv.2021.104988. - DOI - PMC - PubMed
-
- Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, Wang Q, Tan L, Wu W, Tang S, Xiong Z, Zheng S. 2020. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol 58:e00461-20. doi:10.1128/JCM.00461-20. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

