sgDI-tector: defective interfering viral genome bioinformatics for detection of coronavirus subgenomic RNAs
- PMID: 34937774
- PMCID: PMC8848934
- DOI: 10.1261/rna.078969.121
sgDI-tector: defective interfering viral genome bioinformatics for detection of coronavirus subgenomic RNAs
Abstract
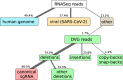
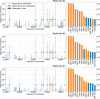
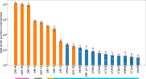
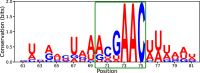
Coronavirus RNA-dependent RNA polymerases produce subgenomic RNAs (sgRNAs) that encode viral structural and accessory proteins. User-friendly bioinformatic tools to detect and quantify sgRNA production are urgently needed to study the growing number of next-generation sequencing (NGS) data of SARS-CoV-2. We introduced sgDI-tector to identify and quantify sgRNA in SARS-CoV-2 NGS data. sgDI-tector allowed detection of sgRNA without initial knowledge of the transcription-regulatory sequences. We produced NGS data and successfully detected the nested set of sgRNAs with the ranking M > ORF3a > N>ORF6 > ORF7a > ORF8 > S > E>ORF7b. We also compared the level of sgRNA production with other types of viral RNA products such as defective interfering viral genomes.
Keywords: SARS-CoV-2; defective viral genomes; subgenomic RNA; user-friendly bioinformatics.
© 2022 Di Gioacchino et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Davidson AD, Williamson MK, Lewis S, Shoemark D, Carroll MW, Heesom KJ, Zambon M, Ellis J, Lewis PA, Hiscox JA, et al. 2020. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med 12: 1–15. 10.1186/s13073-020-00763-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
