A Direct MS-Based Approach to Profile Human Milk Secretory Immunoglobulin A (IgA1) Reveals Donor-Specific Clonal Repertoires With High Longitudinal Stability
- PMID: 34938298
- PMCID: PMC8685336
- DOI: 10.3389/fimmu.2021.789748
A Direct MS-Based Approach to Profile Human Milk Secretory Immunoglobulin A (IgA1) Reveals Donor-Specific Clonal Repertoires With High Longitudinal Stability
Abstract
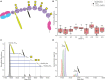
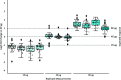
Recently, a mass spectrometry-based approach was introduced to directly assess the IgG1 immunoglobulin clonal repertoires in plasma. Here we expanded upon this approach by describing a mass spectrometry-based technique to assess specifically the clonal repertoire of another important class of immunoglobulin molecules, IgA1, and show it is efficiently and robustly applicable to either milk or plasma samples. Focusing on two individual healthy donors, whose milk was sampled longitudinally during the first 16 weeks of lactation, we demonstrate that the total repertoire of milk sIgA1 is dominated by only 50-500 clones, even though the human body theoretically can generate several orders of magnitude more clones. We show that in each donor the sIgA1 repertoire only changes marginally and quite gradually over the monitored 16-week period of lactation. Furthermore, the observed overlap in clonal repertoires between the two individual donors is close to non-existent. Mothers provide protection to their newborn infants directly by the transfer of antibodies via breastfeeding. The approach introduced here, can be used to visualize the clonal repertoire transferred from mother to infant and to detect changes in-time in that repertoire adapting to changes in maternal physiology.
Keywords: antibody response; antigen binding fragment; clonal repertoire; human milk; mass spectrometry; secretory immunoglobulin A.
Copyright © 2021 Bondt, Dingess, Hoek, van Rijswijck and Heck.
Conflict of interest statement
KD was enrolled as PhD student at Utrecht University during this study and received financial support from Danone Nutricia Research. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

