Recognition of the Possible miRNA-mRNA Controlling Network in Stroke by Bioinformatics Examination
- PMID: 34938355
- PMCID: PMC8687781
- DOI: 10.1155/2021/6745282
Recognition of the Possible miRNA-mRNA Controlling Network in Stroke by Bioinformatics Examination
Retraction in
-
Retracted: Recognition of the Possible miRNA-mRNA Controlling Network in Stroke by Bioinformatics Examination.Comput Math Methods Med. 2023 Dec 6;2023:9828914. doi: 10.1155/2023/9828914. eCollection 2023. Comput Math Methods Med. 2023. PMID: 38094342 Free PMC article.
Abstract
Background: Based on the latest research of WHO, it has been revealed that more than 15 million people suffer from stroke every year worldwide. Of these 15 million people, 6 million succumb to death, and 5 million get permanently disabled. This is the prime reason for the substantial economic burden on all parts of the world.
Methods: These data have been obtained from the GEO database, and the GEO2R tool was used to find out the differentially expressed miRNAs (DEMs) between the stroke and normal patients' blood. FunRich and miRNet were considered to find potential upstream transcription factors and downstream target genes of candidate EMRs. Next, we use GO annotation and KEGG pathway enrichment. Target genes were analyzed with the help of the R software. Then, the STRING database and Cytoscape software were used to conduct PPI and DEM-hub gene networks. Finally, GSE58294 was used to estimate the hub gene expressions.
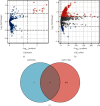
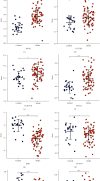
Results: Six DEMs in total were selected out from GSE95204 and GSE117064 datasets. 663 DEMs' target genes were predicted, and NRF1, EGR1, MYC, YY1, E2F1, SP4, and SP1 were predicted as an upstream transcription factor for DEMs' target genes. Target genes of DEMs were primarily augmented in the PI3K-Akt signaling pathway and p53 signaling pathway. The network construction of DEM hygiene is potentially modulated by hsa-miR-3591-5p, hsa-miR-548as-3p, hsa-miR-206, and hsa-miR-4503 hub genes which were found among the top 10 of the hub genes. Among the top 10 hub genes, justification of CTNNB1, PTEN, ESR1, CCND1, KRAS, AKT1, CCND2, CDKN1B, and MYCN was constant with that in the GSE58294 dataset.
Conclusion: In summary, our research first constructs the miRNA-mRNA network in stroke, which probably renders an awakening purview into the pathogenesis and cure of stroke.
Copyright © 2021 Wei Li et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Rothwell P. M., Coull A. J., Silver L. E., et al. Population-based study of event-rate, incidence, case fatality, and mortality for all acute vascular events in all arterial territories (Oxford Vascular Study) Lancet (London, England) . 2005;366(9499):1773–1783. doi: 10.1016/S0140-6736(05)67702-1. - DOI - PubMed
-
- Christiane M., Kirsten L., Ann-Kathrin E., Hussey D. J., Jörg H., Richard H. MicroRNAs: key regulators of chemotherapy response and metastatic potential via complex control of target pathways in esophageal adenocarcinoma. Surgical Oncology . 2018;27(3):392–401. doi: 10.1016/j.suronc.2018.04.001. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

