Antigen presenting cell response to polysaccharide A is characterized by the generation of anti-inflammatory macrophages
- PMID: 34939104
- PMCID: PMC8934142
- DOI: 10.1093/glycob/cwab111
Antigen presenting cell response to polysaccharide A is characterized by the generation of anti-inflammatory macrophages
Abstract
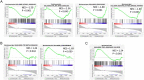
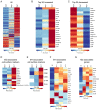
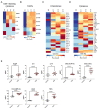
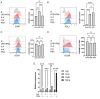
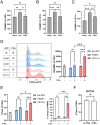
Polysaccharide A (PSA) is the immunodominant capsular carbohydrate from the gram negative commensal microbe Bacteroides fragilis that has shown remarkable potency in ameliorating many rodent models of inflammatory disease by eliciting downstream suppressive CD4+ T cells. PSA is composed of a zwitterionic repeating unit that allows it to be processed by antigen presenting cells (APCs) and presented by MHCII in a glycosylation-dependent manner. While previous work has uncovered much about the interactions between MHCII and PSA, as well as the downstream T cell response, little is known about how PSA affects the phenotype of MHCII+ APCs, including macrophages. Here, we utilized an unbiased systems approach consisting of RNAseq transcriptomics, high-throughput flow cytometry, Luminex analysis and targeted validation experiments to characterize the impact of PSA-mediated stimulation of splenic MHCII+ cells. The data revealed that PSA potently elicited the upregulation of an alternatively activated M2 macrophage transcriptomic and cell surface signature. Cell-type-specific validation experiments further demonstrated that PSA-exposed bone marrow-derived macrophages (BMDMs) induced cell surface and intracellular markers associated with M2 macrophages compared with conventional peptide ovalbumin (ova)-exposed BMDMs. In contrast to macrophages, we also found that CD11c+ dendritic cells (DCs) upregulated the pro-T cell activation costimulatory molecule CD86 following PSA stimulation. Consistent with the divergent BMDM and DC changes, PSA-exposed DCs elicited an antigen-experienced T cell phenotype in co-cultures, whereas macrophages did not. These findings collectively demonstrate that the PSA-induced immune response is characterized by both T cell stimulation via presentation by DCs, and a previously unrecognized anti-inflammatory polarization of macrophages.
Keywords: RNAseq; immune regulation; macrophage; polysaccharide; transcriptomics.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

