Lateral Flow Immunoassay of SARS-CoV-2 Antigen with SERS-Based Registration: Development and Comparison with Traditional Immunoassays
- PMID: 34940267
- PMCID: PMC8699720
- DOI: 10.3390/bios11120510
Lateral Flow Immunoassay of SARS-CoV-2 Antigen with SERS-Based Registration: Development and Comparison with Traditional Immunoassays
Abstract
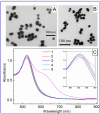
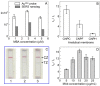
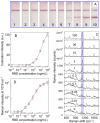
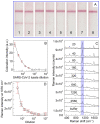
The current COVID-19 pandemic has increased the demand for pathogen detection methods that combine low detection limits with rapid results. Despite the significant progress in methods and devices for nucleic acid amplification, immunochemical methods are still preferred for mass testing without specialized laboratories and highly qualified personnel. The most widely used immunoassays are microplate enzyme-linked immunosorbent assay (ELISA) with photometric detection and lateral flow immunoassay (LFIA) with visual results assessment. However, the disadvantage of ELISA is its considerable duration, and that of LFIA is its low sensitivity. In this study, the modified LFIA of a specific antigen of the causative agent of COVID-19, spike receptor-binding domain, was developed and characterized. This modified LFIA includes the use of gold nanoparticles with immobilized antibodies and 4-mercaptobenzoic acid as surface-enhanced Raman scattering (SERS) nanotag and registration of the nanotag binding by SERS spectrometry. To enhance the sensitivity of LFIA-SERS analysis, we determined the optimal compositions of SERS nanotags and membranes used in LFIA. For benchmark comparison, ELISA and conventional colorimetric LFIA were used with the same immune reagents. The proposed method combines a low detection limit of 0.1 ng/mL (at 0.4 ng/mL for ELISA and 1 ng/mL for qualitative LFIA) with a short assay time equal to 20 min (at 3.5 h for ELISA and 15 min for LFIA). The results obtained demonstrate the promise of using the SERS effects in membrane immuno-analytical systems.
Keywords: Raman spectra; SARS-CoV-2; immunochromatography; surface antigen; test strips.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
-
- Wassie G.T., Azene A.G., Bantie G.M., Dessie G., Aragaw A.M. Incubation period of severe acute respiratory syndrome novel coronavirus 2 that causes coronavirus disease 2019: A systematic review and meta-analysis. Curr. Ther. Res. Clin. Exp. 2020;93:100607. doi: 10.1016/j.curtheres.2020.100607. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

