The Development of SARS-CoV-2 Variants: The Gene Makes the Disease
- PMID: 34940505
- PMCID: PMC8705434
- DOI: 10.3390/jdb9040058
The Development of SARS-CoV-2 Variants: The Gene Makes the Disease
Abstract
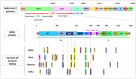
A novel coronavirus (SARS-CoV-2) emerged towards the end of 2019 that caused a severe respiratory disease in humans called COVID-19. It led to a pandemic with a high rate of morbidity and mortality that is ongoing and threatening humankind. Most of the mutations occurring in SARS-CoV-2 are synonymous or deleterious, but a few of them produce improved viral functions. The first known mutation associated with higher transmissibility, D614G, was detected in early 2020. Since then, the virus has evolved; new mutations have occurred, and many variants have been described. Depending on the genes affected and the location of the mutations, they could provide altered infectivity, transmissibility, or immune escape. To date, mutations that cause variations in the SARS-CoV-2 spike protein have been among the most studied because of the protein's role in the initial virus-cell contact and because it is the most variable region in the virus genome. Some concerning mutations associated with an impact on viral fitness have been described in the Spike protein, such as D614G, N501Y, E484K, K417N/T, L452R, and P681R, among others. To understand the impact of the infectivity and antigenicity of the virus, the mutation landscape of SARS-CoV-2 has been under constant global scrutiny. The virus variants are defined according to their origin, their genetic profile (some characteristic mutations prevalent in the lineage), and the severity of the disease they produce, which determines the level of concern. If they increase fitness, new variants can outcompete others in the population. The Alpha variant was more transmissible than previous versions and quickly spread globally. The Beta and Gamma variants accumulated mutations that partially escape the immune defenses and affect the effectiveness of vaccines. Nowadays, the Delta variant, identified around March 2021, has spread and displaced the other variants, becoming the most concerning of all lineages that have emerged. The Delta variant has a particular genetic profile, bearing unique mutations, such as T478K in the spike protein and M203R in the nucleocapsid. This review summarizes the current knowledge of the different mutations that have appeared in SARS-CoV-2, mainly on the spike protein. It analyzes their impact on the protein function and, subsequently, on the level of concern of different variants and their importance in the ongoing pandemic.
Keywords: COVID-19 vaccines; SARS-CoV-2 genome; Spike protein; escape mutation; neutralizing antibodies (nAbs); receptor binding domain (RBD); variant of concern (VOC).
Conflict of interest statement
The author declares no conflict of interest.
Figures
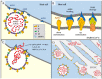
References
-
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and Avian coronaviruses as the gene source of gammacoronavirus and deltacoro-navi. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

