Common Features in lncRNA Annotation and Classification: A Survey
- PMID: 34940758
- PMCID: PMC8708962
- DOI: 10.3390/ncrna7040077
Common Features in lncRNA Annotation and Classification: A Survey
Abstract
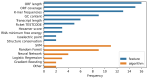
Long non-coding RNAs (lncRNAs) are widely recognized as important regulators of gene expression. Their molecular functions range from miRNA sponging to chromatin-associated mechanisms, leading to effects in disease progression and establishing them as diagnostic and therapeutic targets. Still, only a few representatives of this diverse class of RNAs are well studied, while the vast majority is poorly described beyond the existence of their transcripts. In this review we survey common in silico approaches for lncRNA annotation. We focus on the well-established sets of features used for classification and discuss their specific advantages and weaknesses. While the available tools perform very well for the task of distinguishing coding sequence from other RNAs, we find that current methods are not well suited to distinguish lncRNAs or parts thereof from other non-protein-coding input sequences. We conclude that the distinction of lncRNAs from intronic sequences and untranslated regions of coding mRNAs remains a pressing research gap.
Keywords: classification problems; coding sequence; feature extraction; lncRNA; machine learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

