Understanding CLL biology through mouse models of human genetics
- PMID: 34940815
- PMCID: PMC8703365
- DOI: 10.1182/blood.2021011993
Understanding CLL biology through mouse models of human genetics
Abstract
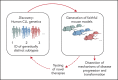
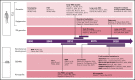
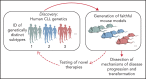
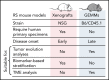
Rapid advances in large-scale next-generation sequencing studies of human samples have progressively defined the highly heterogeneous genetic landscape of chronic lymphocytic leukemia (CLL). At the same time, the numerous challenges posed by the difficulties in rapid manipulation of primary B cells and the paucity of CLL cell lines have limited the ability to interrogate the function of the discovered putative disease "drivers," defined in human sequencing studies through statistical inference. Mouse models represent a powerful tool to study mechanisms of normal and malignant B-cell biology and for preclinical testing of novel therapeutics. Advances in genetic engineering technologies, including the introduction of conditional knockin/knockout strategies, have opened new opportunities to model genetic lesions in a B-cell-restricted context. These new studies build on the experience of generating the MDR mice, the first example of a genetically faithful CLL model, which recapitulates the most common genomic aberration of human CLL: del(13q). In this review, we describe the application of mouse models to the studies of CLL pathogenesis and disease transformation from an indolent to a high-grade malignancy (ie, Richter syndrome [RS]) and treatment, with a focus on newly developed genetically inspired mouse lines modeling recurrent CLL genetic events. We discuss how these novel mouse models, analyzed using new genomic technologies, allow the dissection of mechanisms of disease evolution and response to therapy with greater depth than previously possible and provide important insight into human CLL and RS pathogenesis and therapeutic vulnerabilities. These models thereby provide valuable platforms for functional genomic analyses and treatment studies.
© 2021 by The American Society of Hematology.
Figures


References
-
- Döhner H, Stilgenbauer S, Benner A, et al. . Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910-1916. - PubMed
-
- Quesada V, Conde L, Villamor N, et al. . Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47-52. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

