chinmo-mutant spermatogonial stem cells cause mitotic drive by evicting non-mutant neighbors from the niche
- PMID: 34942115
- PMCID: PMC8752517
- DOI: 10.1016/j.devcel.2021.12.004
chinmo-mutant spermatogonial stem cells cause mitotic drive by evicting non-mutant neighbors from the niche
Abstract
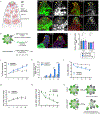
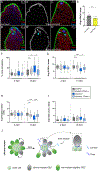
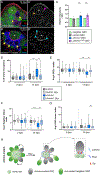
Niches maintain a finite pool of stem cells via restricted space and short-range signals. Stem cells compete for limited niche resources, but the mechanisms regulating competition are poorly understood. Using the Drosophila testis model, we show that germline stem cells (GSCs) lacking the transcription factor Chinmo gain a competitive advantage for niche access. Surprisingly, chinmo-/- GSCs rely on a new mechanism of competition in which they secrete the extracellular matrix protein Perlecan to selectively evict non-mutant GSCs and then upregulate Perlecan-binding proteins to remain in the altered niche. Over time, the GSC pool can be entirely replaced with chinmo-/- cells. As a consequence, the mutant chinmo allele acts as a gene drive element; the majority of offspring inherit the allele despite the heterozygous genotype of the parent. Our results suggest that the influence of GSC competition may extend beyond individual stem cell niche dynamics to population-level allelic drift and evolution.
Keywords: Drosophila; Dystroglycan; Perlecan; aging; biased inheritance; chinmo; competition; germline stem cell; niche; testis.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- BRAND AH & PERRIMON N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

