APC7 mediates ubiquitin signaling in constitutive heterochromatin in the developing mammalian brain
- PMID: 34942119
- PMCID: PMC8741739
- DOI: 10.1016/j.molcel.2021.11.031
APC7 mediates ubiquitin signaling in constitutive heterochromatin in the developing mammalian brain
Abstract
Neurodevelopmental cognitive disorders provide insights into mechanisms of human brain development. Here, we report an intellectual disability syndrome caused by the loss of APC7, a core component of the E3 ubiquitin ligase anaphase promoting complex (APC). In mechanistic studies, we uncover a critical role for APC7 during the recruitment and ubiquitination of APC substrates. In proteomics analyses of the brain from mice harboring the patient-specific APC7 mutation, we identify the chromatin-associated protein Ki-67 as an APC7-dependent substrate of the APC in neurons. Conditional knockout of the APC coactivator protein Cdh1, but not Cdc20, leads to the accumulation of Ki-67 protein in neurons in vivo, suggesting that APC7 is required for the function of Cdh1-APC in the brain. Deregulated neuronal Ki-67 upon APC7 loss localizes predominantly to constitutive heterochromatin. Our findings define an essential function for APC7 and Cdh1-APC in neuronal heterochromatin regulation, with implications for understanding human brain development and disease.
Keywords: APC7; Cdh1; Ki-67; anaphase-promoting complex; brain; chromatin; heterochromatin; neurodevelopment; ubiquitin; ubiquitin ligase.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests. A.B. is an employee of Roche. B.S. is on the scientific advisory board of BioTheryX and Interline Therapeutics, a shareholder of Interline Therapeutics, and a co-inventor of intellectual property licensed to Cinsano.
Figures



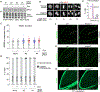

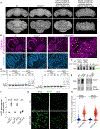

References
-
- Adams DJ, Quail MA, Cox T, Van Der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, et al. (2005). A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86, 753–758. - PubMed
-
- Avagliano L, Parenti I, Grazioli P, Di Fede E, Parodi C, Mariani M, Kaiser FJ, Selicorni A, Gervasini C, and Massa V (2020). Chromatinopathies: A focus on Cornelia de Lange syndrome. Clin. Genet. 97, 3–11. - PubMed
-
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C, Morin GB, Harley CB, Shay JW, Lichtsteiner S, and Wright WE (1998). Extension of Life-Span by Introduction of Telomerase into Normal Human Cells. 279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

