Evolution of enhanced innate immune evasion by SARS-CoV-2
- PMID: 34942634
- PMCID: PMC8850198
- DOI: 10.1038/s41586-021-04352-y
Evolution of enhanced innate immune evasion by SARS-CoV-2
Erratum in
-
Publisher Correction: Evolution of enhanced innate immune evasion by SARS-CoV-2.Nature. 2022 Apr;604(7905):E14. doi: 10.1038/s41586-022-04653-w. Nature. 2022. PMID: 35332335 Free PMC article. No abstract available.
Abstract
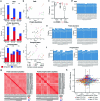
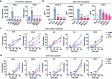
The emergence of SARS-CoV-2 variants of concern suggests viral adaptation to enhance human-to-human transmission1,2. Although much effort has focused on the characterization of changes in the spike protein in variants of concern, mutations outside of spike are likely to contribute to adaptation. Here, using unbiased abundance proteomics, phosphoproteomics, RNA sequencing and viral replication assays, we show that isolates of the Alpha (B.1.1.7) variant3 suppress innate immune responses in airway epithelial cells more effectively than first-wave isolates. We found that the Alpha variant has markedly increased subgenomic RNA and protein levels of the nucleocapsid protein (N), Orf9b and Orf6-all known innate immune antagonists. Expression of Orf9b alone suppressed the innate immune response through interaction with TOM70, a mitochondrial protein that is required for activation of the RNA-sensing adaptor MAVS. Moreover, the activity of Orf9b and its association with TOM70 was regulated by phosphorylation. We propose that more effective innate immune suppression, through enhanced expression of specific viral antagonist proteins, increases the likelihood of successful transmission of the Alpha variant, and may increase in vivo replication and duration of infection4. The importance of mutations outside the spike coding region in the adaptation of SARS-CoV-2 to humans is underscored by the observation that similar mutations exist in the N and Orf9b regulatory regions of the Delta and Omicron variants.
© 2021. The Author(s).
Conflict of interest statement
The N.J.K. laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. N.J.K. has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics and Interline Therapeutics. He is a shareholder in Tenaya Therapeutics, Maze Therapeutics and Interline Therapeutics, has received stocks from Maze Therapeutics and Interline Therapeutics and is a financially compensated Scientific Advisory Board Member for GEn1E Lifesciences. The A.G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold, Model Medicines, Atea Pharma and Merck. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, Paratus and Pfizer. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by the Icahn School of Medicine at Mount Sinai, New York.
Figures






Update of
-
Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant.bioRxiv [Preprint]. 2021 Jun 7:2021.06.06.446826. doi: 10.1101/2021.06.06.446826. bioRxiv. 2021. Update in: Nature. 2022 Feb;602(7897):487-495. doi: 10.1038/s41586-021-04352-y. PMID: 34127972 Free PMC article. Updated. Preprint.
Comment in
-
SARS-CoV-2 learned the 'Alpha'bet of immune evasion.Nat Immunol. 2022 Mar;23(3):351-353. doi: 10.1038/s41590-022-01148-8. Nat Immunol. 2022. PMID: 35194206 No abstract available.
References
-
- Volz E, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 108079/WT_/Wellcome Trust/United Kingdom
- MR/S023380/1/MRC_/Medical Research Council/United Kingdom
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- T32 GM141323/GM/NIGMS NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- F32 CA239333/CA/NCI NIH HHS/United States
- 207511/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- R01 GM133981/GM/NIGMS NIH HHS/United States
- 207511/WT_/Wellcome Trust/United Kingdom
- P01 AI063302/AI/NIAID NIH HHS/United States
- P50 AI150476/AI/NIAID NIH HHS/United States
- R01 AI143292/AI/NIAID NIH HHS/United States
- 206298/WT_/Wellcome Trust/United Kingdom
- U54 CA260560/CA/NCI NIH HHS/United States
- G0801172/MRC_/Medical Research Council/United Kingdom
- 207498/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- 223065/WT_/Wellcome Trust/United Kingdom
- 207498/WT_/Wellcome Trust/United Kingdom
- R01 AI120694/AI/NIAID NIH HHS/United States
- MR/W005611/1/MRC_/Medical Research Council/United Kingdom
- U19 AI135972/AI/NIAID NIH HHS/United States
- 75N93021C00014/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

