Understanding the Role of the Antioxidant Drug Erdosteine and Its Active Metabolite on Staphylococcus aureus Methicillin Resistant Biofilm Formation
- PMID: 34943025
- PMCID: PMC8698571
- DOI: 10.3390/antiox10121922
Understanding the Role of the Antioxidant Drug Erdosteine and Its Active Metabolite on Staphylococcus aureus Methicillin Resistant Biofilm Formation
Abstract
Increasing numbers of researches have suggested that some drugs with reactive oxygen species (ROS)-mediated mechanisms of action modulate biofilm formation of some pathogenic strains. However, the full contribution of ROS to biofilm development is still an open question. In this paper, the correlations between the antioxidant drug Erdosteine (Er) and its active Metabolite I (Met I), ROS and biofilm development of two strains of methicillin resistant Staphylococcus aureus are presented. Experiments revealed that Er and Met I at 2 and 5 mg/L increased up to three orders of magnitude the number of biofilm-dwelling cells, while the content of ROS within the biofilms was reduced above the 87%, with a major effect of Met I in comparison to Er. Comparative proteomics showed that, 5 mg/L Met I modified the expression of 30% and 65% of total proteins in the two strains respectively. Some proteins involved in cell replication were upregulated, and a nitric oxide-based mechanism is assumed to modulate the biofilm development by changing quorum sensitive pathways. Additionally, several proteins involved in virulence were downregulated in the presence of Met I, suggesting that treated cells, despite being greater in number, might have lost part of their virulence.
Keywords: S. aureus; antioxidant; biofilm; erdosteine; metabolite I; nitric oxide; oxidative stress; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
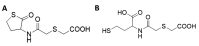





Similar articles
-
[Determination of Biofilm Formation Properties of Methicillin Sensitive and Resistant Staphylococcus aureus Isolates by Conventional and Molecular Methods].Mikrobiyol Bul. 2020 Apr;54(2):223-234. doi: 10.5578/mb.69204. Mikrobiyol Bul. 2020. PMID: 32723278 Turkish.
-
Quantitative NMR metabolite profiling of methicillin-resistant and methicillin-susceptible Staphylococcus aureus discriminates between biofilm and planktonic phenotypes.J Proteome Res. 2014 Jun 6;13(6):2973-85. doi: 10.1021/pr500120c. Epub 2014 May 8. J Proteome Res. 2014. PMID: 24809402 Free PMC article.
-
Methicillin resistance and biofilm production in clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus in México.Biomedica. 2019 Sep 1;39(3):513-523. doi: 10.7705/biomedica.4131. Biomedica. 2019. PMID: 31584765 Free PMC article. English, Spanish.
-
Methicillin-resistant food-related Staphylococcus aureus: a review of current knowledge and biofilm formation for future studies and applications.Res Microbiol. 2017 Jan;168(1):1-15. doi: 10.1016/j.resmic.2016.08.001. Epub 2016 Aug 17. Res Microbiol. 2017. PMID: 27542729 Review.
-
Effect of vancomycin-coated tympanostomy tubes on methicillin-resistant Staphylococcus aureus biofilm formation: in vitro study.J Laryngol Otol. 2010 Jun;124(6):594-8. doi: 10.1017/S0022215109992672. Epub 2010 Jan 8. J Laryngol Otol. 2010. PMID: 20056010 Review.
Cited by
-
The Effects of Mucolytic Agents N-Acetylcysteine and Erdosteine on Hemostasis in Humans.Sovrem Tekhnologii Med. 2024;16(4):55-60. doi: 10.17691/stm2024.16.4.06. Epub 2024 Aug 30. Sovrem Tekhnologii Med. 2024. PMID: 39881835 Free PMC article.
-
A Lactobacilli diet that confers MRSA resistance causes amino acid depletion and increased antioxidant levels in the C. elegans host.Front Microbiol. 2022 Jul 28;13:886206. doi: 10.3389/fmicb.2022.886206. eCollection 2022. Front Microbiol. 2022. PMID: 35966651 Free PMC article.
References
-
- Di Domenico E.G., Rimoldi S.G., Cavallo I., D’Agosto G., Trento E., Cagnoni G., Palazzin A., Pagani C., Romeri F., De Vecchi E., et al. Microbial biofilm correlates with an increased antibiotic tolerance and poor therapeutic outcome in infective endocarditis. BMC Microbiol. 2019;19:228. doi: 10.1186/s12866-019-1596-2. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

