The Adaptor Protein Lurap1 Is Required for Cell Cohesion during Epiboly Movement in Zebrafish
- PMID: 34943252
- PMCID: PMC8699034
- DOI: 10.3390/biology10121337
The Adaptor Protein Lurap1 Is Required for Cell Cohesion during Epiboly Movement in Zebrafish
Abstract
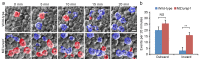
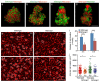
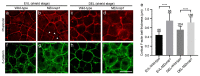
Cell adhesion and polarized cellular behaviors play critical roles in a wide variety of morphogenetic events. In the zebrafish embryo, epiboly represents an important process of epithelial morphogenesis that involves differential cell adhesion and dynamic cell shape changes for coordinated movements of different cell populations, but the underlying mechanism remains poorly understood. The adaptor protein Lurap1 functions to link myotonic dystrophy kinase-related Rac/Cdc42-binding kinase with MYO18A for actomyosin retrograde flow in cell migration. We previously reported that it interacts with Dishevelled in convergence and extension movements during gastrulation. Here, we show that it regulates blastoderm cell adhesion and radial cell intercalation during epiboly. In zebrafish mutant embryos with loss of both maternal and zygotic Lurap1 function, deep cell multilayer of the blastoderm exhibit delayed epiboly with respect to the superficial layer. Time-lapse imaging reveals that these deep cells undergo unstable intercalation, which impedes their expansion over the yolk cell. Cell sorting and adhesion assays indicate reduced cellular cohesion of the blastoderm. These defects are correlated with disrupted cytoskeletal organization in the cortex of blastoderm cells. Thus, the present results extend our previous works by demonstrating that Lurap1 is required for cell adhesion and cell behavior changes to coordinate cell movements during epithelial morphogenesis. They provide insights for a further understanding of the regulation of cytoskeletal organization during gastrulation cell movements.
Keywords: F-actin; Lurap1; cell adhesion; cytoskeleton; epiboly; gastrulation; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

