Mutational Diversity in the Quinolone Resistance-Determining Regions of Type-II Topoisomerases of Salmonella Serovars
- PMID: 34943668
- PMCID: PMC8698434
- DOI: 10.3390/antibiotics10121455
Mutational Diversity in the Quinolone Resistance-Determining Regions of Type-II Topoisomerases of Salmonella Serovars
Abstract
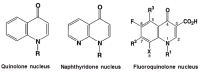
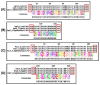
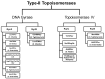
Quinolone resistance in bacterial pathogens has primarily been associated with mutations in the quinolone resistance-determining regions (QRDRs) of bacterial type-II topoisomerases, which are DNA gyrase and topoisomerase IV. Depending on the position and type of the mutation (s) in the QRDRs, bacteria either become partially or completely resistant to quinolone. QRDR mutations have been identified and characterized in Salmonella enterica isolates from around the globe, particularly during the last decade, and efforts have been made to understand the propensity of different serovars to carry such mutations. Because there is currently no thorough analysis of the available literature on QRDR mutations in different Salmonella serovars, this review aims to provide a comprehensive picture of the mutational diversity in QRDRs of Salmonella serovars, summarizing the literature related to both typhoidal and non-typhoidal Salmonella serovars with a special emphasis on recent findings. This review will also discuss plasmid-mediated quinolone-resistance determinants with respect to their additive or synergistic contributions with QRDR mutations in imparting elevated quinolone resistance. Finally, the review will assess the contribution of membrane transporter-mediated quinolone efflux to quinolone resistance in strains carrying QRDR mutations. This information should be helpful to guide the routine surveillance of foodborne Salmonella serovars, especially with respect to their spread across countries, as well as to improve laboratory diagnosis of quinolone-resistant Salmonella strains.
Keywords: DNA gyrase; Salmonella; fluoroquinolones; food-borne pathogens; non-typhoidal Salmonella (NTS); plasmid-mediated quinolone resistance (PMQR); quinolone resistance-determining regions (QRDRs); topoisomerase IV; typhoidal Salmonella.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Presence of plasmid-mediated quinolone resistance (PMQR) genes in non-typhoidal Salmonella strains with reduced susceptibility to fluoroquinolones isolated from human salmonellosis in Gyeonggi-do, South Korea from 2016 to 2019.Gut Pathog. 2021 Jun 1;13(1):35. doi: 10.1186/s13099-021-00431-7. Gut Pathog. 2021. PMID: 34074336 Free PMC article.
-
Presence of plasmid-mediated quinolone resistance determinants and mutations in gyrase and topoisomerase in Salmonella enterica isolates with resistance and reduced susceptibility to ciprofloxacin.Diagn Microbiol Infect Dis. 2016 May;85(1):85-9. doi: 10.1016/j.diagmicrobio.2016.01.016. Epub 2016 Jan 26. Diagn Microbiol Infect Dis. 2016. PMID: 26971183
-
Quinolone Resistance Mechanisms Among Salmonella enterica in Malaysia.Microb Drug Resist. 2016 Jun;22(4):259-72. doi: 10.1089/mdr.2015.0158. Epub 2015 Dec 18. Microb Drug Resist. 2016. PMID: 26683630
-
Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control.Foods. 2018 Oct 11;7(10):167. doi: 10.3390/foods7100167. Foods. 2018. PMID: 30314348 Free PMC article. Review.
-
The Rise of Non-typhoidal Salmonella Infections in India: Causes, Symptoms, and Prevention.Cureus. 2023 Oct 8;15(10):e46699. doi: 10.7759/cureus.46699. eCollection 2023 Oct. Cureus. 2023. PMID: 38021876 Free PMC article. Review.
Cited by
-
Prophage Gifsy-1 Induction in Salmonella enterica Serovar Typhimurium Reduces Persister Cell Formation after Ciprofloxacin Exposure.Microbiol Spectr. 2023 Aug 17;11(4):e0187423. doi: 10.1128/spectrum.01874-23. Epub 2023 Jun 12. Microbiol Spectr. 2023. PMID: 37306609 Free PMC article.
-
Ciprofloxacin resistance and bile-induced biofilm enhancement in Salmonella paratyphi A isolates.New Microbes New Infect. 2025 Jun 2;66:101602. doi: 10.1016/j.nmni.2025.101602. eCollection 2025 Aug. New Microbes New Infect. 2025. PMID: 40688374 Free PMC article.
-
Synthesis and antibacterial activity evaluation of N (7) position-modified balofloxacins.Front Chem. 2022 Aug 19;10:963442. doi: 10.3389/fchem.2022.963442. eCollection 2022. Front Chem. 2022. PMID: 36059868 Free PMC article.
-
A Longitudinal Study on the Dynamics of Salmonella enterica Prevalence and Serovar Composition in Beef Cattle Feces and Lymph Nodes and Potential Contributing Sources from the Feedlot Environment.Appl Environ Microbiol. 2023 Apr 26;89(4):e0003323. doi: 10.1128/aem.00033-23. Epub 2023 Apr 6. Appl Environ Microbiol. 2023. PMID: 37022263 Free PMC article.
-
Antimicrobial resistance in Salmonella: One Health perspective on global food safety challenges.Sci One Health. 2025 Jun 28;4:100117. doi: 10.1016/j.soh.2025.100117. eCollection 2025. Sci One Health. 2025. PMID: 40687400 Free PMC article. Review.
References
-
- Brighty K.E., Gootz T.D. The Quinolones. 3rd ed. Academic Press; Cambridge, MA, USA: 2000. Chemistry and mechanism of action of the quinolone antibacterials.
-
- Hamer D.H., Gorbach S.L. The Quinolones. Academic Press; Cambridge, MA, USA: 2000. Use of the quinolones for treatment and prophylaxis of bacterial gastrointestinal infections; p. 303.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

