Optimized 3D Culture of Hepatic Cells for Liver Organoid Metabolic Assays
- PMID: 34943788
- PMCID: PMC8699701
- DOI: 10.3390/cells10123280
Optimized 3D Culture of Hepatic Cells for Liver Organoid Metabolic Assays
Abstract
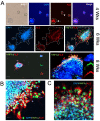
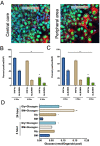
The liver is among the principal organs for glucose homeostasis and metabolism. Studies of liver metabolism are limited by the inability to expand primary hepatocytes in vitro while maintaining their metabolic functions. Human hepatic three-dimensional (3D) organoids have been established using defined factors, yet hepatic organoids from adult donors showed impaired expansion. We examined conditions to facilitate the expansion of adult donor-derived hepatic organoids (HepAOs) and HepG2 cells in organoid cultures (HepGOs) using combinations of growth factors and small molecules. The expansion dynamics, gluconeogenic and HNF4α expression, and albumin secretion are assessed. The conditions tested allow the generation of HepAOs and HepGOs in 3D cultures. Nevertheless, gluconeogenic gene expression varies greatly between conditions. The organoid expansion rates are limited when including the TGFβ inhibitor A8301, while are relatively higher with Forskolin (FSK) and Oncostatin M (OSM). Notably, expanded HepGOs grown in the optimized condition maintain detectable gluconeogenic expression in a spatiotemporal distribution at 8 weeks. We present optimized conditions by limiting A8301 and incorporating FSK and OSM to allow the expansion of HepAOs from adult donors and HepGOs with gluconeogenic competence. These models increase the repertoire of human hepatic cellular tools available for use in liver metabolic assays.
Keywords: 3D culture; HepG2 cells; gluconeogenesis; liver regeneration; metabolic assays; organoids.
Conflict of interest statement
Rutgers University has filed a provisional patent application on the methods described in the manuscript. H.E.S. is the scientific founder of Celvive, Inc. (New Brunswick, NJ, USA).
Figures




References
-
- Alexander M., Loomis A.K., van der Lei J., Duarte-Salles T., Prieto-Alhambra D., Ansell D., Pasqua A., Lapi F., Rijnbeek P., Mosseveld M., et al. Risks and clinical predictors of cirrhosis and hepatocellular carcinoma diagnoses in adults with diagnosed NAFLD: Real-world study of 18 million patients in four European cohorts. BMC Med. 2019;17:95. doi: 10.1186/s12916-019-1321-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

