Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi
- PMID: 34944084
- PMCID: PMC8700503
- DOI: 10.3390/cells10123574
Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi
Abstract
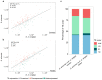
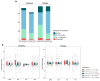
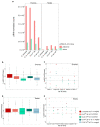
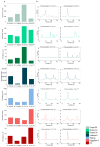
Interspecific hybridization may lead to sterility and/or inviability through differential expression of genes and transposable elements (TEs). In Drosophila, studies have reported massive TE mobilization in hybrids from interspecific crosses of species presenting high divergence times. However, few studies have examined the consequences of TE mobilization upon hybridization in recently diverged species, such as Drosophila arizonae and D. mojavensis. We have sequenced transcriptomes of D. arizonae and the subspecies D. m. wrigleyi and their reciprocal hybrids, as well as piRNAs, to analyze the impact of genomic stress on TE regulation. Our results revealed that the differential expression in both gonadal tissues of parental species was similar. Globally, ovaries and testes showed few deregulated TEs compared with both parental lines. Analyses of small RNA data showed that in ovaries, the TE upregulation is likely due to divergence of copies inherited from parental genomes and lack of piRNAs mapping to them. Nevertheless, in testes, the divergent expression of genes associated with chromatin state and piRNA pathway potentially indicates that TE differential expression is related to the divergence of regulatory genes that play a role in modulating transcriptional and post-transcriptional mechanisms.
Keywords: expression; hybrids; repleta group; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

