p66α Suppresses Breast Cancer Cell Growth and Migration by Acting as Co-Activator of p53
- PMID: 34944103
- PMCID: PMC8700327
- DOI: 10.3390/cells10123593
p66α Suppresses Breast Cancer Cell Growth and Migration by Acting as Co-Activator of p53
Abstract
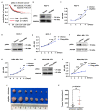
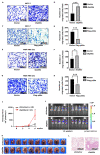
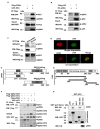
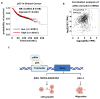
p66α is a GATA zinc finger domain-containing transcription factor that has been shown to be essential for gene silencing by participating in the NuRD complex. Several studies have suggested that p66α is a risk gene for a wide spectrum of diseases such as diabetes, schizophrenia, and breast cancer; however, its biological role has not been defined. Here, we report that p66α functions as a tumor suppressor to inhibit breast cancer cell growth and migration, evidenced by the fact that the depletion of p66α results in accelerated tumor growth and migration of breast cancer cells. Mechanistically, immunoprecipitation assays identify p66α as a p53-interacting protein that binds the DNA-binding domain of p53 molecule predominantly via its CR2 domain. Depletion of p66α in multiple breast cells results in decreased expression of p53 target genes, while over-expression of p66α results in increased expression of these target genes. Moreover, p66α promotes the transactivity of p53 by enhancing p53 binding at target promoters. Together, these findings demonstrate that p66α is a tumor suppressor by functioning as a co-activator of p53.
Keywords: breast cancer; cell growth; metastasis; migration; p53; p66α.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Loss of TRIM31 promotes breast cancer progression through regulating K48- and K63-linked ubiquitination of p53.Cell Death Dis. 2021 Oct 14;12(10):945. doi: 10.1038/s41419-021-04208-3. Cell Death Dis. 2021. PMID: 34650049 Free PMC article.
-
FXR deficiency induced ferroptosis via modulation of the CBP-dependent p53 acetylation to suppress breast cancer growth and metastasis.Cell Death Dis. 2024 Nov 14;15(11):826. doi: 10.1038/s41419-024-07222-3. Cell Death Dis. 2024. PMID: 39543094 Free PMC article.
-
Hotspot mutant p53-R273H inhibits KLF6 expression to promote cell migration and tumor metastasis.Cell Death Dis. 2020 Jul 30;11(7):595. doi: 10.1038/s41419-020-02814-1. Cell Death Dis. 2020. PMID: 32733026 Free PMC article.
-
PRR11 and SKA2 gene pair is overexpressed and regulated by p53 in breast cancer.BMB Rep. 2019 Feb;52(2):157-162. doi: 10.5483/BMBRep.2019.52.2.207. BMB Rep. 2019. PMID: 30760381 Free PMC article.
-
Unique features of the anti-parallel, heterodimeric coiled-coil interaction between methyl-cytosine binding domain 2 (MBD2) homologues and GATA zinc finger domain containing 2A (GATAD2A/p66α).J Biol Chem. 2013 Feb 1;288(5):3419-27. doi: 10.1074/jbc.M112.431346. Epub 2012 Dec 13. J Biol Chem. 2013. PMID: 23239876 Free PMC article.
Cited by
-
What Genetic Modifications of Source Pigs Are Essential and Sufficient for Cell, Tissue, and Organ Xenotransplantation?Transpl Int. 2024 Dec 4;37:13681. doi: 10.3389/ti.2024.13681. eCollection 2024. Transpl Int. 2024. PMID: 39697899 Free PMC article. Review.
-
DLC1 inhibits colon adenocarcinoma cell migration by promoting secretion of the neurotrophic factor MANF.Front Oncol. 2022 Sep 14;12:900166. doi: 10.3389/fonc.2022.900166. eCollection 2022. Front Oncol. 2022. PMID: 36185184 Free PMC article.
-
Formation of Re-Aggregated Neonatal Porcine Islet Clusters Improves In Vitro Function and Transplantation Outcome.Transpl Int. 2022 Dec 22;35:10697. doi: 10.3389/ti.2022.10697. eCollection 2022. Transpl Int. 2022. PMID: 36685665 Free PMC article.
-
Deep Proteomics Analysis Unravels the Molecular Signatures of Tonsillar B Cells in PFAPA and OSAS in the Pediatric Population.Int J Mol Sci. 2025 Jul 10;26(14):6621. doi: 10.3390/ijms26146621. Int J Mol Sci. 2025. PMID: 40724870 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

