EmbedDTI: Enhancing the Molecular Representations via Sequence Embedding and Graph Convolutional Network for the Prediction of Drug-Target Interaction
- PMID: 34944427
- PMCID: PMC8698792
- DOI: 10.3390/biom11121783
EmbedDTI: Enhancing the Molecular Representations via Sequence Embedding and Graph Convolutional Network for the Prediction of Drug-Target Interaction
Abstract
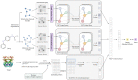
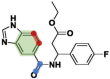
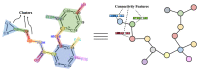
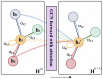
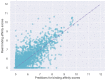
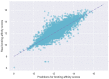
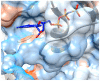
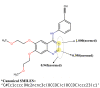
The identification of drug-target interaction (DTI) plays a key role in drug discovery and development. Benefitting from large-scale drug databases and verified DTI relationships, a lot of machine-learning methods have been developed to predict DTIs. However, due to the difficulty in extracting useful information from molecules, the performance of these methods is limited by the representation of drugs and target proteins. This study proposes a new model called EmbedDTI to enhance the representation of both drugs and target proteins, and improve the performance of DTI prediction. For protein sequences, we leverage language modeling for pretraining the feature embeddings of amino acids and feed them to a convolutional neural network model for further representation learning. For drugs, we build two levels of graphs to represent compound structural information, namely the atom graph and substructure graph, and adopt graph convolutional network with an attention module to learn the embedding vectors for the graphs. We compare EmbedDTI with the existing DTI predictors on two benchmark datasets. The experimental results show that EmbedDTI outperforms the state-of-the-art models, and the attention module can identify the components crucial for DTIs in compounds.
Keywords: drug-target interaction; graph convolutional network; molecular representation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Heifetz A., Southey M., Morao I., Townsend-Nicholson A. Computational Methods Used in Hit-to-Lead and Lead Optimization Stages of Structure-Based Drug Discovery. Methods Mol. Biol. 2018;1705:375–394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

