Exploring the Value of BRD9 as a Biomarker, Therapeutic Target and Co-Target in Prostate Cancer
- PMID: 34944438
- PMCID: PMC8698755
- DOI: 10.3390/biom11121794
Exploring the Value of BRD9 as a Biomarker, Therapeutic Target and Co-Target in Prostate Cancer
Abstract
Background and aims: Despite recent advances in advanced prostate cancer treatments, clinical biomarkers or treatments for men with such cancers are imperfect. Targeted therapies have shown promise, but there remain fewer actionable targets in prostate cancer than in other cancers. This work aims to characterise BRD9, currently understudied in prostate cancer, and investigate its co-expression with other genes to assess its potential as a biomarker and therapeutic target in human prostate cancer.
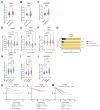
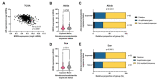
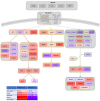
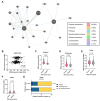
Materials and methods: Omics data from a total of 2053 prostate cancer patients across 11 independent datasets were accessed via Cancertool and cBioPortal. mRNA M.expression and co-expression, mutations, amplifications, and deletions were assessed with respect to key clinical parameters including survival, Gleason grade, stage, progression, and treatment. Network and pathway analysis was carried out using Genemania, and heatmaps were constructed using Morpheus.
Results: BRD9 is overexpressed in prostate cancer patients, especially those with metastatic disease. BRD9 expression did not differ in patients treated with second generation antiandrogens versus those who were not. BRD9 is co-expressed with many genes in the SWI/SNF and BET complexes, as well as those in common signalling pathways in prostate cancer.
Summary and conclusions: BRD9 has potential as a diagnostic and prognostic biomarker in prostate cancer. BRD9 also shows promise as a therapeutic target, particularly in advanced prostate cancer, and as a co-target alongside other genes in the SWI/SNF and BET complexes, and those in common prostate cancer signalling pathways. These promising results highlight the need for wider experimental inhibition and co-targeted inhibition of BRD9 in vitro and in vivo, to build on the limited inhibition data available.
Keywords: BRD9; SWI/SNF; prostate cancer; targeted therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Prostate Cancer UK About Prostate Cancer. [(accessed on 19 April 2021)]. Available online: https://prostatecanceruk.org/prostate-information/about-prostate-cancer.
-
- Prostate Cancer UK Prostate Cancer UK’s Best Practice Pathway TREATMENT. Prostate Cancer UK, 2019. [(accessed on 19 April 2021)]. Available online: https://prostatecanceruk.org/media/1fknrpz5/treatmentpathwaypathwaycomme....
-
- NICE Information about PSA Testing. [(accessed on 19 April 2021)]. Available online: https://cks.nice.org.uk/topics/prostate-cancer/diagnosis/psa-testing/#:~....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

