Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data
- PMID: 34944830
- PMCID: PMC8699503
- DOI: 10.3390/cancers13246210
Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data
Abstract
Background: Immune checkpoint inhibitors (ICIs) are now a therapeutic standard in advanced non-small cell lung cancer (NSCLC), but strong predictive markers for ICIs efficacy are still lacking. We evaluated machine learning models built on simple clinical and biological data to individually predict response to ICIs.
Methods: Patients with metastatic NSCLC who received ICI in second line or later were included. We collected clinical and hematological data and studied the association of this data with disease control rate (DCR), progression free survival (PFS) and overall survival (OS). Multiple machine learning (ML) algorithms were assessed for their ability to predict response.
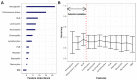
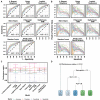
Results: Overall, 298 patients were enrolled. The overall response rate and DCR were 15.3% and 53%, respectively. Median PFS and OS were 3.3 and 11.4 months, respectively. In multivariable analysis, DCR was significantly associated with performance status (PS) and hemoglobin level (OR 0.58, p < 0.0001; OR 1.8, p < 0.001). These variables were also associated with PFS and OS and ranked top in random forest-based feature importance. Neutrophil-to-lymphocyte ratio was also associated with DCR, PFS and OS. The best ML algorithm was a random forest. It could predict DCR with satisfactory efficacy based on these three variables. Ten-fold cross-validated performances were: accuracy 0.68 ± 0.04, sensitivity 0.58 ± 0.08; specificity 0.78 ± 0.06; positive predictive value 0.70 ± 0.08; negative predictive value 0.68 ± 0.06; AUC 0.74 ± 0.03.
Conclusion: Combination of simple clinical and biological data could accurately predict disease control rate at the individual level.
Keywords: blood counts; lung cancer; machine learning; prediction; response; survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Planchard D., Popat S., Kerr K., Novello S., Smit E.F., Faivre-Finn C., Mok T.S., Reck M., Van Schil P.E., Hellmann M.D., et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018;29((Suppl. 4)):iv192–iv237. doi: 10.1093/annonc/mdy275. - DOI - PubMed
-
- Hopkins A.M., Rowland A., Kichenadasse G., Wiese M.D., Gurney H., McKinnon R.A., Karapetis C.S., Sorich M.J. Predicting response and toxicity to immune checkpoint inhibitors using routinely available blood and clinical markers. Br. J. Cancer. 2017;117:913–920. doi: 10.1038/bjc.2017.274. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

