Increased Replication Stress Determines ATR Inhibitor Sensitivity in Neuroblastoma Cells
- PMID: 34944835
- PMCID: PMC8699051
- DOI: 10.3390/cancers13246215
Increased Replication Stress Determines ATR Inhibitor Sensitivity in Neuroblastoma Cells
Abstract
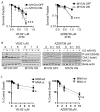
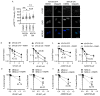
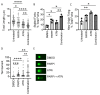
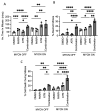
Despite intensive high-dose multimodal therapy, high-risk neuroblastoma (NB) confers a less than 50% survival rate. This study investigates the role of replication stress in sensitivity to inhibition of Ataxia telangiectasia and Rad3-related (ATR) in pre-clinical models of high-risk NB. Amplification of the oncogene MYCN always imparts high-risk disease and occurs in 25% of all NB. Here, we show that MYCN-induced replication stress directly increases sensitivity to the ATR inhibitors VE-821 and AZD6738. PARP inhibition with Olaparib also results in replication stress and ATR activation, and sensitises NB cells to ATR inhibition independently of MYCN status, with synergistic levels of cell death seen in MYCN expressing ATR- and PARP-inhibited cells. Mechanistically, we demonstrate that ATR inhibition increases the number of persistent stalled and collapsed replication forks, exacerbating replication stress. It also abrogates S and G2 cell cycle checkpoints leading to death during mitosis in cells treated with an ATR inhibitor combined with PARP inhibition. In summary, increased replication stress through high MYCN expression, PARP inhibition or chemotherapeutic agents results in sensitivity to ATR inhibition. Our findings provide a mechanistic rationale for the inclusion of ATR and PARP inhibitors as a potential treatment strategy for high-risk NB.
Keywords: ATR; MYCN; PARP; neuroblastoma; replication stress.
Conflict of interest statement
N.J.C. has received research funding from Aguoron Pharmaceuticals and Pfizer for the development of rucaparib, BioMarin for studies on talazoparib and Tesaro for studies with niraparib and paid consultancy from Abbvie for veliparib. Newcastle University receives royalty income from the sales of rucaparib that are shared with contributors to its development, including N.J.C., N.J.C. has also received research funding from Vertex Pharmaceuticals and is currently in receipt of funding from Merck KGaA for work on ATR inhibitors and holds a patent for a pharmacodynamic biomarker of ATR inhibition (WO2014055756A1). All other authors declare no conflict of interest.
Figures



References
-
- Cohn S.L., Pearson A.D.J., London W.B., Monclair T., Ambros P.F., Brodeur G.M., Faldum A., Hero B., Iehara T., Machin D., et al. The international neuroblastoma risk group (INRG) classification system: An INRG task force report. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. - DOI - PMC - PubMed
-
- Basta N.O., Halliday G.C., Makin G., Birch J., Feltbower R., Bown N., Elliott M., Moreno L., Barone G., Pearson A.D., et al. Factors associated with recurrence and survival length following relapse in patients with neuroblastoma. Br. J. Cancer. 2016;115:1048–1057. doi: 10.1038/bjc.2016.302. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

