hsa_circ_0001275 Is One of a Number of circRNAs Dysregulated in Enzalutamide Resistant Prostate Cancer and Confers Enzalutamide Resistance In Vitro
- PMID: 34945002
- PMCID: PMC8715667
- DOI: 10.3390/cancers13246383
hsa_circ_0001275 Is One of a Number of circRNAs Dysregulated in Enzalutamide Resistant Prostate Cancer and Confers Enzalutamide Resistance In Vitro
Abstract
Background: Enzalutamide is part of the treatment regimen for metastatic castration-resistant prostate cancer (MCRPC). However, both intrinsic and acquired resistance to the drug remain substantial clinical quandaries. circRNAs, a novel type of non-coding RNA, have been identified in a number of cancers including prostate cancer and have been associated with cancer development and progression. circRNAs have shown great potential as clinically useful blood-based 'liquid biopsies' and as therapeutic targets in prostate cancer. The aim of this study was to examine the role of circRNA transcripts in enzalutamide-resistant prostate cancer cells and assess their utility as biomarkers.
Methods: An isogenic cell line model of enzalutamide resistance was subjected to circRNA microarray profiling. Several differentially expressed circRNAs, along with their putative parental genes were validated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). circRNAs of interest were stably overexpressed in the control cell line and drug sensitivity was assessed using an ELISA-based proliferation assay. The candidate circRNA, hsa_circ_0001275, was measured in patient plasma samples using RT-droplet digital PCR (RT-ddPCR).
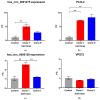
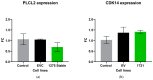
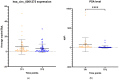
Results: hsa_circ_0001275 and its parental gene, PLCL2, were significantly up-regulated in strongly resistant clones vs. control (p < 0.05). Overexpression of hsa_circ_0001275 in the control cell line resulted in increased resistance to enzalutamide (p < 0.05). While RT-ddPCR analysis of hsa_circ_0001275 expression in plasma samples of 44 clinical trial participants showed a trend that mirrored the stages of disease activity (as defined by PSA level), the association did not reach statistical significance.
Conclusions: Our data suggest that increased levels of hsa_circ_0001275 contribute to enzalutamide resistance. hsa_circ_0001275 plasma expression showed a trend that mirrors the PSA level at specific disease time points, indicating that circRNAs mirror disease recurrence and burden and may be associated with enzalutamide resistance.
Keywords: circRNA (circular RNA); enzalutamide resistance; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






References
-
- Noone A.M., Howlader N., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., et al. SEER Cancer Statistics Review 1975–2015. National Cancer Institute; Bethesda, MD, USA: Apr, 2018.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

