Comparative Analysis of Salmon Cell Lines and Zebrafish Primary Cell Cultures Infection with the Fish Pathogen Piscirickettsia salmonis
- PMID: 34946119
- PMCID: PMC8706985
- DOI: 10.3390/microorganisms9122516
Comparative Analysis of Salmon Cell Lines and Zebrafish Primary Cell Cultures Infection with the Fish Pathogen Piscirickettsia salmonis
Abstract
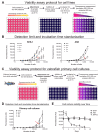
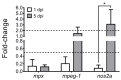
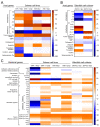
Piscirickettsia salmonis is the etiologic agent of piscirickettsiosis, a disease that causes significant losses in the salmon farming industry. In order to unveil the pathogenic mechanisms of P. salmonis, appropriate molecular and cellular studies in multiple cell lines with different origins need to be conducted. Toward that end, we established a cell viability assay that is suitable for high-throughput analysis using the alamarBlue reagent to follow the distinct stages of the bacterial infection cycle. Changes in host cell viability can be easily detected using either an absorbance- or fluorescence-based plate reader. Our method accurately tracked the infection cycle across two different Atlantic salmon-derived cell lines, with macrophage and epithelial cell properties, and zebrafish primary cell cultures. Analyses were also carried out to quantify intracellular bacterial replication in combination with fluorescence microscopy to visualize P. salmonis and cellular structures in fixed cells. In addition, dual gene expression analysis showed that the pro-inflammatory cytokines IL-6, IL-12, and TNFα were upregulated, while the cytokines IL1b and IFNγ were downregulated in the three cell culture types. The expression of the P. salmonis metal uptake and heme acquisition genes, together with the toxin and effector genes ospD3, ymt, pipB2 and pepO, were upregulated at the early and late stages of infection regardless of the cell culture type. On the other hand, Dot/Icm secretion system genes as well as stationary state and nutrient scarcity-related genes were upregulated only at the late stage of P. salmonis intracellular infection. We propose that these genes encoding putative P. salmonis virulence factors and immune-related proteins could be suitable biomarkers of P. salmonis infection. The infection protocol and cell viability assay described here provide a reliable method to compare the molecular and cellular changes induced by P. salmonis in other cell lines and has the potential to be used for high-throughput screenings of novel antimicrobials targeting this important fish intracellular pathogen.
Keywords: P. salmonis virulence; cell culture viability; host–pathogen interaction; infection biomarkers; kidney primary cell culture; salmon cell lines; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Transcriptomic Changes of Piscirickettsia salmonis During Intracellular Growth in a Salmon Macrophage-Like Cell Line.Front Cell Infect Microbiol. 2020 Jan 9;9:426. doi: 10.3389/fcimb.2019.00426. eCollection 2019. Front Cell Infect Microbiol. 2020. PMID: 31998656 Free PMC article.
-
Global Proteomic Profiling of Piscirickettsia salmonis and Salmon Macrophage-Like Cells during Intracellular Infection.Microorganisms. 2020 Nov 24;8(12):1845. doi: 10.3390/microorganisms8121845. Microorganisms. 2020. PMID: 33255149 Free PMC article.
-
Transcriptional response of Atlantic salmon families to Piscirickettsia salmonis infection highlights the relevance of the iron-deprivation defence system.BMC Genomics. 2015 Jul 4;16(1):495. doi: 10.1186/s12864-015-1716-9. BMC Genomics. 2015. PMID: 26141111 Free PMC article.
-
Piscirickettsiosis and Piscirickettsia salmonis in fish: a review.J Fish Dis. 2014 Mar;37(3):163-88. doi: 10.1111/jfd.12211. Epub 2013 Nov 26. J Fish Dis. 2014. PMID: 24279295 Review.
-
Vaccines for piscirickettsiosis (salmonid rickettsial septicaemia, SRS): the Chile perspective.Expert Rev Vaccines. 2017 Mar;16(3):215-228. doi: 10.1080/14760584.2017.1244483. Epub 2016 Oct 12. Expert Rev Vaccines. 2017. PMID: 27690686 Review.
Cited by
-
Collective behavior and virulence arsenal of the fish pathogen Piscirickettsia salmonis in the biofilm realm.Front Cell Infect Microbiol. 2022 Dec 5;12:1067514. doi: 10.3389/fcimb.2022.1067514. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36544910 Free PMC article.
-
Immunonutrition: facilitating mucosal immune response in teleost intestine with amino acids through oxidant-antioxidant balance.Front Immunol. 2023 Sep 29;14:1241615. doi: 10.3389/fimmu.2023.1241615. eCollection 2023. Front Immunol. 2023. PMID: 37841275 Free PMC article. Review.
References
-
- Almendras F.E., Fuentealba I.C. Salmonid rickettsial septicemia caused by Piscirickettsia salmonis: A review. Dis. Aquat. Organ. 1997;29:137–144. doi: 10.3354/dao029137. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

