Identification and Characterization of a Novel Recurrent ERCC6 Variant in Patients with a Severe Form of Cockayne Syndrome B
- PMID: 34946871
- PMCID: PMC8701866
- DOI: 10.3390/genes12121922
Identification and Characterization of a Novel Recurrent ERCC6 Variant in Patients with a Severe Form of Cockayne Syndrome B
Abstract
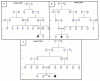
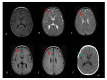
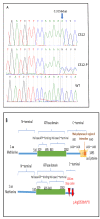
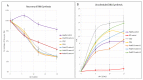
Cockayne syndrome (CS) is a rare disease caused by mutations in ERCC6/CSB or ERCC8/CSA. We report here the clinical, genetic, and functional analyses of three unrelated patients mutated in ERCC6/CSB with a severe phenotype. After clinical examination, two patients were investigated via next generation sequencing, targeting seventeen Nucleotide Excision Repair (NER) genes. All three patients harbored a novel, c.3156dup, homozygous mutation located in exon 18 of ERCC6/CSB that affects the C-terminal region of the protein. Sanger sequencing confirmed the mutation and the parental segregation in the three families, and Western blots showed a lack of the full-length protein. NER functional impairment was shown by reduced recovery of RNA synthesis with proficient unscheduled DNA synthesis after UV-C radiations in patient-derived fibroblasts. Despite sharing the same mutation, the clinical spectrum was heterogeneous among the three patients, and only two patients displayed clinical photosensitivity. This novel ERCC6 variant in Tunisian patients suggests a founder effect and has implications for setting-up prenatal diagnosis/genetic counselling in North Africa, where this disease is largely undiagnosed. This study reveals one of the rare cases of CS clinical heterogeneity despite the same mutation. Moreover, the occurrence of an identical homozygous mutation, which either results in clinical photosensitivity or does not, strongly suggests that this classic CS symptom relies on multiple factors.
Keywords: Cockayne syndrome; DNA repair disorder; ERCC6; accelerated aging; neurodegeneration.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures

Similar articles
-
Heterogeneous clinical features in Cockayne syndrome patients and siblings carrying the same CSA mutations.Orphanet J Rare Dis. 2022 Mar 5;17(1):121. doi: 10.1186/s13023-022-02257-1. Orphanet J Rare Dis. 2022. PMID: 35248096 Free PMC article.
-
Functional and clinical relevance of novel mutations in a large cohort of patients with Cockayne syndrome.J Med Genet. 2018 May;55(5):329-343. doi: 10.1136/jmedgenet-2017-104877. Epub 2018 Mar 23. J Med Genet. 2018. PMID: 29572252
-
First molecular study in Lebanese patients with Cockayne syndrome and report of a novel mutation in ERCC8 gene.BMC Med Genet. 2018 Sep 10;19(1):161. doi: 10.1186/s12881-018-0677-7. BMC Med Genet. 2018. PMID: 30200888 Free PMC article.
-
Novel frame shift mutation in ERCC6 leads to a severe form of Cockayne syndrome with postnatal growth failure and early death: A case report and brief literature review.Medicine (Baltimore). 2018 Aug;97(33):e11636. doi: 10.1097/MD.0000000000011636. Medicine (Baltimore). 2018. PMID: 30113454 Free PMC article. Review.
-
Cockayne Syndrome Group B (CSB): The Regulatory Framework Governing the Multifunctional Protein and Its Plausible Role in Cancer.Cells. 2021 Apr 10;10(4):866. doi: 10.3390/cells10040866. Cells. 2021. PMID: 33920220 Free PMC article. Review.
Cited by
-
Immunity in the Progeroid Model of Cockayne Syndrome: Biomarkers of Pathological Aging.Cells. 2024 Feb 26;13(5):402. doi: 10.3390/cells13050402. Cells. 2024. PMID: 38474366 Free PMC article.
-
Heterogeneous clinical features in Cockayne syndrome patients and siblings carrying the same CSA mutations.Orphanet J Rare Dis. 2022 Mar 5;17(1):121. doi: 10.1186/s13023-022-02257-1. Orphanet J Rare Dis. 2022. PMID: 35248096 Free PMC article.
-
Supplementation with nicotinamide limits accelerated aging in affected individuals with cockayne syndrome and restores antioxidant defenses.Aging (Albany NY). 2024 Nov 26;16(21):13271-13287. doi: 10.18632/aging.206160. Epub 2024 Nov 26. Aging (Albany NY). 2024. PMID: 39611850 Free PMC article.
-
A matter of delicate balance: Loss and gain of Cockayne syndrome proteins in premature aging and cancer.Front Aging. 2022 Jul 21;3:960662. doi: 10.3389/fragi.2022.960662. eCollection 2022. Front Aging. 2022. PMID: 35935726 Free PMC article. Review.
-
Insights Into Cockayne Syndrome Type B: What Underlies Its Pathogenesis?Aging Cell. 2025 Jul;24(7):e70136. doi: 10.1111/acel.70136. Epub 2025 Jun 19. Aging Cell. 2025. PMID: 40536083 Free PMC article. Review.
References
-
- Calmels N., Botta E., Jia N., Fawcett H., Nardo T., Nakazawa Y., Lanzafame M., Moriwaki S., Sugita K., Kubota M., et al. Functional and clinical relevance of novel mutations in a large cohort of patients with Cockayne syndrome. J. Med. Genet. 2018;55:329–343. doi: 10.1136/jmedgenet-2017-104877. - DOI - PubMed
-
- Wilson B.T., Stark Z., Sutton R.E., Danda S., Ekbote A.V., Elsayed S.M., Gibson L., Goodship J.A., Jackson A.P., Keng W.T., et al. The Cockayne Syndrome Natural History (CoSyNH) study: Clinical findings in 102 individuals and recommendations for care. Genet. Med. 2016;18:483–493. doi: 10.1038/gim.2015.110. - DOI - PMC - PubMed
-
- Fisher A., Asghar M., Ryan S., Lynch B., Green A., Knerr I. GP59 A rare cause of ‘mitochondrial disorder’: Cockayne syndrome. Arch. Dis. Child. 2019;104:A53. doi: 10.1136/archdischild-2019-epa.125. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

