Optical Genome Mapping in Routine Human Genetic Diagnostics-Its Advantages and Limitations
- PMID: 34946907
- PMCID: PMC8701374
- DOI: 10.3390/genes12121958
Optical Genome Mapping in Routine Human Genetic Diagnostics-Its Advantages and Limitations
Abstract
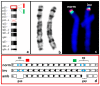
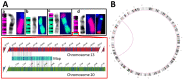
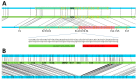
In recent years, optical genome mapping (OGM) has developed into a highly promising method of detecting large-scale structural variants in human genomes. It is capable of detecting structural variants considered difficult to detect by other current methods. Hence, it promises to be feasible as a first-line diagnostic tool, permitting insight into a new realm of previously unknown variants. However, due to its novelty, little experience with OGM is available to infer best practices for its application or to clarify which features cannot be detected. In this study, we used the Saphyr system (Bionano Genomics, San Diego, CA, USA), to explore its capabilities in human genetic diagnostics. To this end, we tested 14 DNA samples to confirm a total of 14 different structural or numerical chromosomal variants originally detected by other means, namely, deletions, duplications, inversions, trisomies, and a translocation. Overall, 12 variants could be confirmed; one deletion and one inversion could not. The prerequisites for detection of similar variants were explored by reviewing the OGM data of 54 samples analyzed in our laboratory. Limitations, some owing to the novelty of the method and some inherent to it, were described. Finally, we tested the successful application of OGM in routine diagnostics and described some of the challenges that merit consideration when utilizing OGM as a diagnostic tool.
Keywords: OGM; cytogenetics; cytogenomics; numerical chromosomal aberrations; optical genome mapping; routine genetic testing; structural chromosomal aberrations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tyson C., Harvard C., Locker R., Friedman J.M., Langlois S., Lewis M.E., Van Allen M., Somerville M., Arbour L., Clarke L., et al. Submicroscopic deletions and duplications in individuals with intellectual disability detected by array-CGH. Am. J. Med. Genet A. 2005;139:173–185. doi: 10.1002/ajmg.a.31015. - DOI - PubMed
-
- Ishihara T., Kikuchi Y., Sandberg A.A. Chromosomes of twenty cancer effusions: Correlation of karyotypic, clinical, and pathologic aspects. J. Natl. Cancer Inst. 1963;30:1303–1361. - PubMed
-
- Bednar M. DNA microarray technology and application. Med. Sci. Monit. 2000;6:796–800. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

