New Mutations in HFE2 and TFR2 Genes Causing Non HFE-Related Hereditary Hemochromatosis
- PMID: 34946929
- PMCID: PMC8702017
- DOI: 10.3390/genes12121980
New Mutations in HFE2 and TFR2 Genes Causing Non HFE-Related Hereditary Hemochromatosis
Abstract
Hereditary hemochromatosis (HH) is an iron metabolism disease clinically characterized by excessive iron deposition in parenchymal organs such as liver, heart, pancreas, and joints. It is caused by mutations in at least five different genes. HFE hemochromatosis is the most common type of hemochromatosis, while non-HFE related hemochromatosis are rare cases. Here, we describe six new patients of non-HFE related HH from five different families. Two families (Family 1 and 2) have novel nonsense mutations in the HFE2 gene have novel nonsense mutations (p.Arg63Ter and Asp36ThrfsTer96). Three families have mutations in the TFR2 gene, one case has one previously unreported mutation (Family A-p.Asp680Tyr) and two cases have known pathogenic mutations (Family B and D-p.Trp781Ter and p.Gln672Ter respectively). Clinical, biochemical, and genetic data are discussed in all these cases. These rare cases of non-HFE related hereditary hemochromatosis highlight the importance of an earlier molecular diagnosis in a specialized center to prevent serious clinical complications.
Keywords: HFE related hereditary hemochromatosis; HFE2 gene; TFR2 gene; homozygous; iron overload; missense; non-HFE related hereditary hemochromatosis; nonsense.
Conflict of interest statement
C.T. is CEO of BloodGenetics, S.L. and M.S. are co-founders of BloodGenetics.
Figures







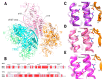
References
-
- Silvestri L., Nai A., Dulja A., Pagani A. Hepcidin and the BMP-SMAD pathway: An unexpected liaison. Vitam. Horm. 2019;110:71–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RTI-2018-101735-B-I100 (MCI/AEI/FEDER, EU)/Spanish Ministry of Science and Innovation (MICINN)
- APU and ADISCON Patient associations
- RTC2019-007074-1/Spanish Ministry of Science and Innovation (MICINN)
- MARIE SKLODOWSKA-CURIE GRANT AGREEMENT NO 894737/European Union’s Horizon 2020 research and innovation programme
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

