Single Cell Sequencing of Induced Pluripotent Stem Cell Derived Retinal Ganglion Cells (iPSC-RGC) Reveals Distinct Molecular Signatures and RGC Subtypes
- PMID: 34946963
- PMCID: PMC8702079
- DOI: 10.3390/genes12122015
Single Cell Sequencing of Induced Pluripotent Stem Cell Derived Retinal Ganglion Cells (iPSC-RGC) Reveals Distinct Molecular Signatures and RGC Subtypes
Abstract
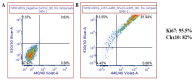
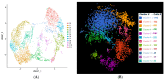
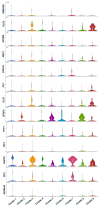
We intend to identify marker genes with differential gene expression (DEG) and RGC subtypes in cultures of human-induced pluripotent stem cell (iPSC)-derived retinal ganglion cells. Single-cell sequencing was performed on mature and functional iPSC-RGCs at day 40 using Chromium Single Cell 3' V3 protocols (10X Genomics). Sequencing libraries were run on Illumina Novaseq to generate 150 PE reads. Demultiplexed FASTQ files were mapped to the hg38 reference genome using the STAR package, and cluster analyses were performed using a cell ranger and BBrowser2 software. QC analysis was performed by removing the reads corresponding to ribosomal and mitochondrial genes, as well as cells that had less than 1X mean absolute deviation (MAD), resulting in 4705 cells that were used for further analyses. Cells were separated into clusters based on the gene expression normalization via PCA and TSNE analyses using the Seurat tool and/or Louvain clustering when using BBrowser2 software. DEG analysis identified subsets of RGCs with markers like MAP2, RBPMS, TUJ1, BRN3A, SOX4, TUBB3, SNCG, PAX6 and NRN1 in iPSC-RGCs. Differential expression analysis between separate clusters identified significant DEG transcripts associated with cell cycle, neuron regulatory networks, protein kinases, calcium signaling, growth factor hormones, and homeobox transcription factors. Further cluster refinement identified RGC diversity and subtype specification within iPSC-RGCs. DEGs can be used as biomarkers for RGC subtype classification, which will allow screening model systems that represent a spectrum of diseases with RGC pathology.
Keywords: FACS analysis; RGC subtypes; clustering; glaucoma; iPSC-RGCs; iPSCs; marker genes; retinal ganglion cells; single cell sequencing; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Dietze J., Blair K., Havens S.J. Glaucoma. StatPearls Publishing; Treasure Island, FL, USA: 2021.
-
- Chavali V.R.M., Haider N., Rathi S., Vrathasha V., Alapati T., He J., Gill K., Nikonov R., Duong T.T., McDougald D.S., et al. Dual SMAD inhibition and Wnt inhibition enable efficient and reproducible differentiations of induced pluripotent stem cells into retinal ganglion cells. Sci. Rep. 2020;10:11828. doi: 10.1038/s41598-020-68811-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

