Origin of Increased Solvent Accessibility of Peptide Bonds in Mutual Synergetic Folding Proteins
- PMID: 34948202
- PMCID: PMC8704591
- DOI: 10.3390/ijms222413404
Origin of Increased Solvent Accessibility of Peptide Bonds in Mutual Synergetic Folding Proteins
Abstract
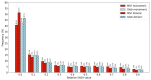
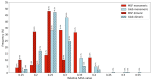
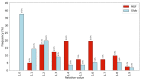
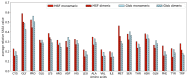
Mutual Synergetic Folding (MSF) proteins belong to a recently discovered class of proteins. These proteins are disordered in their monomeric but ordered in their oligomeric forms. Their amino acid composition is more similar to globular proteins than to disordered ones. Our preceding work shed light on important structural aspects of the structural organization of these proteins, but the background of this behavior is still unknown. We suggest that solvent accessibility is an important factor, especially solvent accessibility of the peptide bonds can be accounted for this phenomenon. The side chains of the amino acids which form a peptide bond have a high local contribution to the shielding of the peptide bond from the solvent. During the oligomerization step, other non-local residues contribute to the shielding. We investigated these local and non-local effects of shielding based on Shannon information entropy calculations. We found that MSF and globular homodimeric proteins have different local contributions resulting from different amino acid pair frequencies. Their non-local distribution is also different because of distinctive inter-subunit contacts.
Keywords: Shannon information entropy; amino acid composition; inter-subunit interaction; intrinsically disordered proteins; mutual synergetic folding; solvent accessibility of peptide bonds; solvent-accessible surface area.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
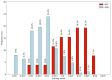

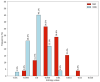
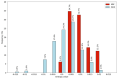
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

