The Carbohydrate Metabolism of Lactiplantibacillus plantarum
- PMID: 34948249
- PMCID: PMC8704671
- DOI: 10.3390/ijms222413452
The Carbohydrate Metabolism of Lactiplantibacillus plantarum
Abstract
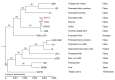
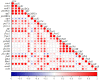
Lactiplantibacillus plantarum has a strong carbohydrate utilization ability. This characteristic plays an important role in its gastrointestinal tract colonization and probiotic effects. L. plantarum LP-F1 presents a high carbohydrate utilization capacity. The genome analysis of 165 L. plantarum strains indicated the species has a plenty of carbohydrate metabolism genes, presenting a strain specificity. Furthermore, two-component systems (TCSs) analysis revealed that the species has more TCSs than other lactic acid bacteria, and the distribution of TCS also shows the strain specificity. In order to clarify the sugar metabolism mechanism under different carbohydrate fermentation conditions, the expressions of 27 carbohydrate metabolism genes, catabolite control protein A (CcpA) gene ccpA, and TCSs genes were analyzed by quantitative real-time PCR technology. The correlation analysis between the expressions of regulatory genes and sugar metabolism genes showed that some regulatory genes were correlated with most of the sugar metabolism genes, suggesting that some TCSs might be involved in the regulation of sugar metabolism.
Keywords: Lactiplantibacillus plantarum; carbohydrate metabolism; two-component system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Turpin W., Weiman M., Guyot J.P., Lajus A., Cruveiller S., Humblot C. The genomic and transcriptomic basis of the potential of Lactobacillus plantarum A6 to improve the nutritional quality of a cereal based fermented food. Int. J. Food Microbiol. 2018;266:346–354. doi: 10.1016/j.ijfoodmicro.2017.10.011. - DOI - PubMed
-
- Kleerebezem M., Boekhorst J., van Kranenburg R., Molenaar D., Kuipers O.P., Leer R., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

