FlyWire: online community for whole-brain connectomics
- PMID: 34949809
- PMCID: PMC8903166
- DOI: 10.1038/s41592-021-01330-0
FlyWire: online community for whole-brain connectomics
Abstract
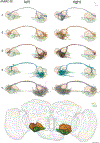
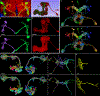
Due to advances in automated image acquisition and analysis, whole-brain connectomes with 100,000 or more neurons are on the horizon. Proofreading of whole-brain automated reconstructions will require many person-years of effort, due to the huge volumes of data involved. Here we present FlyWire, an online community for proofreading neural circuits in a Drosophila melanogaster brain and explain how its computational and social structures are organized to scale up to whole-brain connectomics. Browser-based three-dimensional interactive segmentation by collaborative editing of a spatially chunked supervoxel graph makes it possible to distribute proofreading to individuals located virtually anywhere in the world. Information in the edit history is programmatically accessible for a variety of uses such as estimating proofreading accuracy or building incentive systems. An open community accelerates proofreading by recruiting more participants and accelerates scientific discovery by requiring information sharing. We demonstrate how FlyWire enables circuit analysis by reconstructing and analyzing the connectome of mechanosensory neurons.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
TM and HSS are owners of Zetta AI LLC, which provides neural circuit reconstruction services for research labs. RL and NK are employees of Zetta AI LLC.
Figures







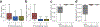

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

